3ZIF
 
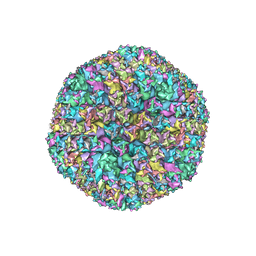 | | Cryo-EM structures of two intermediates provide insight into adenovirus assembly and disassembly | | Descriptor: | HEXON PROTEIN, PENTON PROTEIN, PIX, ... | | Authors: | Cheng, L, Huang, X, Li, X, Xiong, W, Sun, W, Yang, C, Zhang, K, Wang, Y, Liu, H, Ji, G, Sun, F, Zheng, C, Zhu, P. | | Deposit date: | 2013-01-09 | | Release date: | 2014-01-22 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-Em Structures of Two Bovine Adenovirus Type 3 Intermediates
Virology, 450, 2014
|
|
1VM3
 
 | |
1VM5
 
 | |
3QQ4
 
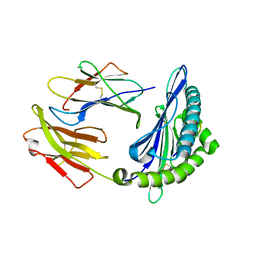 | | Crystal structure of swine major histocompatibility complex class I SLA-1 0401 and identification of 2009 pandemic swine-origin influenza A H1N1 virus cytotoxic T lymphocyte epitope peptides | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, VP35 | | Authors: | Zhang, N, Qi, J, Gao, F, Pan, X, Chen, R, Li, Q, Chen, Z, Li, X, Xia, C, Gao, G.F. | | Deposit date: | 2011-02-15 | | Release date: | 2011-12-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Crystal structure of swine major histocompatibility complex class I SLA-1 0401 and identification of 2009 pandemic swine-origin influenza A H1N1 virus cytotoxic T lymphocyte epitope peptides.
J.Virol., 85, 2011
|
|
8DJM
 
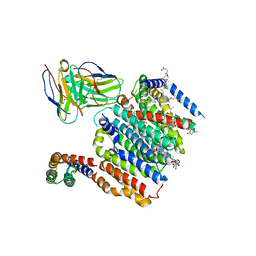 | | HMGCR-UBIAD1 Complex State 1 | | Descriptor: | 3-hydroxy-3-methylglutaryl-coenzyme A reductase, CHOLESTEROL HEMISUCCINATE, Digitonin, ... | | Authors: | Chen, H, Qi, X, Li, X. | | Deposit date: | 2022-07-01 | | Release date: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Regulated degradation of HMG CoA reductase requires conformational changes in sterol-sensing domain.
Nat Commun, 13, 2022
|
|
8DJK
 
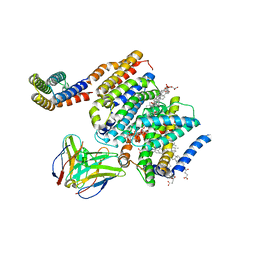 | | HMGCR-UBIAD1 Complex State 2 | | Descriptor: | 3-hydroxy-3-methylglutaryl-coenzyme A reductase, CHOLESTEROL HEMISUCCINATE, Digitonin, ... | | Authors: | Chen, H, Qi, X, Li, X. | | Deposit date: | 2022-06-30 | | Release date: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Regulated degradation of HMG CoA reductase requires conformational changes in sterol-sensing domain.
Nat Commun, 13, 2022
|
|
5LME
 
 | | Specific-DNA binding activity of the cross-brace zinc finger motif of the piggyBac transposase | | Descriptor: | ZINC ION, piggyBac transposase | | Authors: | Morellet, N, Taylor, J.A, Wieninger, S, Moriau, S, Li, X, Lescop, E, Mathy, N, Bischerour, J, Betermier, M, Bardiaux, B, Nilges, M, Craig, N.L, Hickman, A.B, Dyda, F, Guittet, E. | | Deposit date: | 2016-07-30 | | Release date: | 2017-12-20 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Sequence-specific DNA binding activity of the cross-brace zinc finger motif of the piggyBac transposase.
Nucleic Acids Res., 46, 2018
|
|
4MY5
 
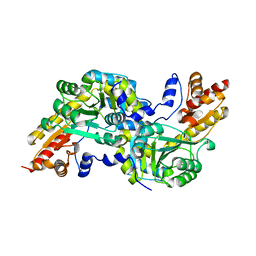 | | Crystal structure of the aromatic amino acid aminotransferase from Streptococcus mutants | | Descriptor: | Putative amino acid aminotransferase | | Authors: | Cong, X, Li, X, Ge, J, Feng, Y, Feng, X, Li, S. | | Deposit date: | 2013-09-27 | | Release date: | 2014-10-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | Crystal structure of the aromatic-amino-acid aminotransferase from Streptococcus mutans.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6XBM
 
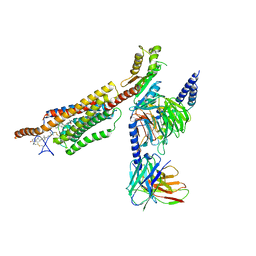 | | Structure of human SMO-Gi complex with 24(S),25-EC | | Descriptor: | 17-[3-(3,3-DIMETHYL-OXIRANYL)-1-METHYL-PROPYL]-10,13-DIMETHYL-2,3,4,7,8,9,10,11,12,13,14,15,16,17-TETRADECAHYDRO-1H-CYC LOPENTA[A]PHENANTHREN-3-OL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Qi, X, Long, T, Li, X. | | Deposit date: | 2020-06-06 | | Release date: | 2020-09-30 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Sterols in an intramolecular channel of Smoothened mediate Hedgehog signaling.
Nat.Chem.Biol., 16, 2020
|
|
6XBL
 
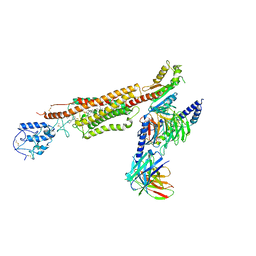 | | Structure of human SMO-Gi complex with SAG | | Descriptor: | 3-chloro-N-[trans-4-(methylamino)cyclohexyl]-N-{[3-(pyridin-4-yl)phenyl]methyl}-1-benzothiophene-2-carboxamide, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Qi, X, Li, X. | | Deposit date: | 2020-06-06 | | Release date: | 2020-09-30 | | Last modified: | 2020-11-25 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Sterols in an intramolecular channel of Smoothened mediate Hedgehog signaling.
Nat.Chem.Biol., 16, 2020
|
|
6XBK
 
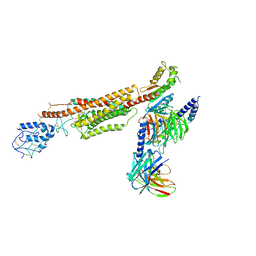 | | Structure of human SMO-G111C/I496C complex with Gi | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Qi, X, Long, T, Li, X. | | Deposit date: | 2020-06-06 | | Release date: | 2020-09-30 | | Last modified: | 2020-11-25 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Sterols in an intramolecular channel of Smoothened mediate Hedgehog signaling.
Nat.Chem.Biol., 16, 2020
|
|
6XBJ
 
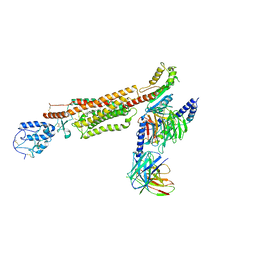 | | Structure of human SMO-D384R complex with Gi | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Qi, X, Long, T, Li, X. | | Deposit date: | 2020-06-06 | | Release date: | 2020-09-30 | | Last modified: | 2020-11-25 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Sterols in an intramolecular channel of Smoothened mediate Hedgehog signaling.
Nat.Chem.Biol., 16, 2020
|
|
7SHN
 
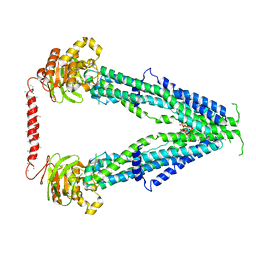 | | Cryo-EM structure of oleoyl-CoA-bound human peroxisomal fatty acid transporter ABCD1 | | Descriptor: | ATP-binding cassette sub-family D member 1, S-{(3R,5R,9R)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-3,5-dioxido-10,14-dioxo-2,4,6-trioxa-11,15-diaza-3lambda~5~,5lambda~5~-diphosphaheptadecan-17-yl} (9Z)-octadec-9-enethioate (non-preferred name) | | Authors: | Wang, R, Li, X. | | Deposit date: | 2021-10-09 | | Release date: | 2021-11-03 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of acyl-CoA transport across the peroxisomal membrane by human ABCD1.
Cell Res., 32, 2022
|
|
7SHM
 
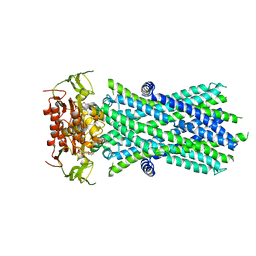 | |
4Q2V
 
 | | Crystal Structure of Ricin A chain complexed with Baicalin inhibitor | | Descriptor: | 5,6-dihydroxy-4-oxo-2-phenyl-4H-chromen-7-yl beta-D-glucopyranosiduronic acid, Ricin | | Authors: | Deng, X, Li, X, Dong, J, Chen, Y. | | Deposit date: | 2014-04-10 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Baicalin inhibits the lethality of ricin in mice by inducing protein oligomerization.
J.Biol.Chem., 290, 2015
|
|
8FNI
 
 | | Cryo-EM structure of RNase-treated RESC-B in trypanosomal RNA editing | | Descriptor: | RNA-editing substrate-binding complex protein 10 (RESC10), RNA-editing substrate-binding complex protein 11 (RESC11), RNA-editing substrate-binding complex protein 13 (RESC13), ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
8FNK
 
 | | Cryo-EM structure of RNase-untreated RESC-B in trypanosomal RNA editing | | Descriptor: | RNA-editing substrate-binding complex protein 10 (RESC10), RNA-editing substrate-binding complex protein 11 (RESC11), RNA-editing substrate-binding complex protein 13 (RESC13), ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
8FNC
 
 | | Cryo-EM structure of RNase-treated RESC-C in trypanosomal RNA editing | | Descriptor: | Mitochondrial RNA binding complex 1 subunit, Mitochondrial RNA binding protein, Phytanoyl-CoA dioxygenase family protein, ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
8FN4
 
 | | Cryo-EM structure of RNase-treated RESC-A in trypanosomal RNA editing | | Descriptor: | RNA-editing substrate-binding complex protein 1 (RESC1), RNA-editing substrate-binding complex protein 2 (RESC2), RNA-editing substrate-binding complex protein 3 (RESC3), ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-26 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
8FN6
 
 | | Cryo-EM structure of RNase-untreated RESC-A in trypanosomal RNA editing | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, RNA-editing substrate-binding complex protein 1 (RESC1), RNA-editing substrate-binding complex protein 2 (RESC2), ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
8FNF
 
 | | Cryo-EM structure of RNase-untreated RESC-C in trypanosomal RNA editing | | Descriptor: | Mitochondrial RNA binding complex 1 subunit, Mitochondrial RNA binding protein, Phytanoyl-CoA dioxygenase family protein, ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
3V34
 
 | | Crystal structure of MCPIP1 conserved domain with magnesium ion in the catalytic center | | Descriptor: | MAGNESIUM ION, Ribonuclease ZC3H12A | | Authors: | Xu, J, Peng, W, Sun, Y, Wang, X, Xu, Y, Li, X, Gao, G, Rao, Z. | | Deposit date: | 2011-12-12 | | Release date: | 2012-05-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Structural study of MCPIP1 N-terminal conserved domain reveals a PIN-like RNase
Nucleic Acids Res., 40, 2012
|
|
3V33
 
 | | Crystal structure of MCPIP1 conserved domain with zinc-finger motif | | Descriptor: | Ribonuclease ZC3H12A | | Authors: | Xu, J, Peng, W, Sun, Y, Wang, X, Xu, Y, Li, X, Gao, G, Rao, Z. | | Deposit date: | 2011-12-12 | | Release date: | 2012-05-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Structural study of MCPIP1 N-terminal conserved domain reveals a PIN-like RNase
Nucleic Acids Res., 40, 2012
|
|
3V32
 
 | | Crystal structure of MCPIP1 N-terminal conserved domain | | Descriptor: | Ribonuclease ZC3H12A | | Authors: | Xu, J, Peng, W, Sun, Y, Wang, X, Xu, Y, Li, X, Gao, G, Rao, Z. | | Deposit date: | 2011-12-12 | | Release date: | 2012-05-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural study of MCPIP1 N-terminal conserved domain reveals a PIN-like RNase
Nucleic Acids Res., 40, 2012
|
|
5C4W
 
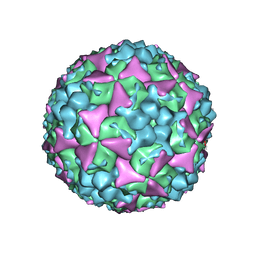 | | Crystal structure of coxsackievirus A16 | | Descriptor: | CHLORIDE ION, POTASSIUM ION, SODIUM ION, ... | | Authors: | Ren, J, Wang, X, Zhu, L, Hu, Z, Gao, Q, Yang, P, Li, X, Wang, J, Shen, X, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2015-06-18 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structures of Coxsackievirus A16 Capsids with Native Antigenicity: Implications for Particle Expansion, Receptor Binding, and Immunogenicity.
J.Virol., 89, 2015
|
|
