3RT2
 
 | | Crystal structure of apo-PYL10 | | Descriptor: | Abscisic acid receptor PYL10 | | Authors: | Hao, Q, Yin, P, Li, W, Wang, L, Yan, C, Wang, J, Yan, N. | | Deposit date: | 2011-05-02 | | Release date: | 2011-06-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Molecular Basis of ABA-Independent Inhibition of PP2Cs by a Subclass of PYL Proteins
Mol.Cell, 42, 2011
|
|
3RQR
 
 | | Crystal structure of the RYR domain of the rabbit ryanodine receptor | | Descriptor: | (UNK)(UNK)(UNK)(UNK), Ryanodine receptor 1 | | Authors: | Nair, U.B, Li, W, Dong, A, Walker, J.R, Gramolini, A, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-04-28 | | Release date: | 2011-06-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural determination of the phosphorylation domain of the ryanodine receptor.
Febs J., 279, 2012
|
|
6KJH
 
 | | Functional and structural insights into the unusual oxyanion hole-like geometry in macrolactin acyltransferase selective for dicarboxylic acyl donors | | Descriptor: | Putative beta-lactamase, SULFATE ION | | Authors: | Xiao, F, Sheng, D, Feng, Y, Li, W. | | Deposit date: | 2019-07-22 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural Basis of Specificity for Carboxyl-Terminated Acyl Donors in a Bacterial Acyltransferase.
J.Am.Chem.Soc., 142, 2020
|
|
6KJQ
 
 | | Functional and structural insights into the unusual oxyanion hole-like geometry in macrolactin acyltransferase selective for dicarboxylic acyl donors | | Descriptor: | (3~{Z},5~{E},8~{S},9~{E},11~{E},14~{S},16~{R},17~{Z},19~{E},24~{R})-24-methyl-8,14,16-tris(oxidanyl)-1-oxacyclotetracosa-3,5,9,11,17,19-hexaen-2-one, Putative beta-lactamase | | Authors: | Xiao, F, Dong, S, Feng, Y, Li, W. | | Deposit date: | 2019-07-23 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis of Specificity for Carboxyl-Terminated Acyl Donors in a Bacterial Acyltransferase.
J.Am.Chem.Soc., 142, 2020
|
|
7XA3
 
 | | Cryo-EM structure of the CCL2 bound CCR2-Gi complex | | Descriptor: | C-C motif chemokine 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Shao, Z, Tan, Y, Shen, Q, Yao, B, Hou, L, Qin, J, Xu, P, Mao, C, Chen, L, Zhang, H, Shen, D, Zhang, C, Li, W, Du, X, Li, F, Chen, Z, Jiang, Y, Xu, H.E, Ying, S, Ma, H, Zhang, Y, Shen, H. | | Deposit date: | 2022-03-17 | | Release date: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular insights into ligand recognition and activation of chemokine receptors CCR2 and CCR3.
Cell Discov, 8, 2022
|
|
7X9Y
 
 | | Cryo-EM structure of the apo CCR3-Gi complex | | Descriptor: | C-C chemokine receptor type 3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Shao, Z, Tan, Y, Shen, Q, Yao, B, Hou, L, Qin, J, Xu, P, Mao, C, Chen, L, Zhang, H, Shen, D, Zhang, C, Li, W, Du, X, Li, F, Chen, Z, Jiang, Y, Xu, H.E, Ying, S, Ma, H, Zhang, Y, Shen, H. | | Deposit date: | 2022-03-16 | | Release date: | 2022-08-24 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular insights into ligand recognition and activation of chemokine receptors CCR2 and CCR3.
Cell Discov, 8, 2022
|
|
7E4Q
 
 | | Crystal structure of tubulin in complex with L-DM1-SMe | | Descriptor: | (1S,2R,3R,5R,6R,16E,18E,20R,21S)-11-chloro-21-hydroxy-12,20-dimethoxy-2,5,9,16-tetramethyl-8,23-dioxo-4,24-dioxa-9,22-diazatetracyclo[19.3.1.1~10,14~.0~3,5~]hexacosa-10(26),11,13,16,18-pentaen-6-yl (2S)-2-{methyl[3-(methyldisulfanyl)propanoyl]amino}propanoate (non-preferred name), 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wang, Y, Li, W. | | Deposit date: | 2021-02-14 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | C3 ester side chain plays a pivotal role in the antitumor activity of Maytansinoids.
Biochem.Biophys.Res.Commun., 566, 2021
|
|
7E4R
 
 | | Crystal structure of tubulin in complex with D-DM1-SMe | | Descriptor: | (10E,12E)-86-chloro-14-hydroxy-85,14-dimethoxy-33,2,7,10-tetramethyl-12,6-dioxo-7-aza-1(6,4)-oxazinana-3(2,3)-oxirana-8(1,3)-benzenacyclotetradecaphane-10,12-dien-4-yl N-methyl-N-(3-(methylsulfinothioyl)propanoyl)-D-alaninate, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wang, Y, Li, W. | | Deposit date: | 2021-02-15 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.597 Å) | | Cite: | C3 ester side chain plays a pivotal role in the antitumor activity of Maytansinoids.
Biochem.Biophys.Res.Commun., 566, 2021
|
|
7EN3
 
 | | Crystal structure of tubulin in complex with Tubulysin analogue TGL | | Descriptor: | (2S,4R)-5-(4-fluorophenyl)-2-methyl-4-[[2-[(1R,3R)-4-methyl-3-[5-methylhexyl-[(2S,3S)-3-methyl-2-[[(2R)-1-methylpiperidin-2-yl]carbonylamino]pentanoyl]amino]-1-oxidanyl-pentyl]-1,3-thiazol-4-yl]carbonylamino]pentanoic acid, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wang, Y, Li, W. | | Deposit date: | 2021-04-15 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.643 Å) | | Cite: | The X-ray structure of tubulysin analogue TGL in complex with tubulin and three possible routes for the development of next-generation tubulysin analogues.
Biochem.Biophys.Res.Commun., 565, 2021
|
|
7E4Z
 
 | | Crystal structure of tubulin in complex with Maytansinol | | Descriptor: | (1R,2S,3S,5S,6S,16E,18E,20R,21S)-11-chloro-6,21-dihydroxy-12,20-dimethoxy-2,5,9,16-tetramethyl-4,24-dioxa-9,22-diazatetracyclo[19.3.1.1~10,14~.0~3,5~]hexacosa-10(26),11,13,16,18-pentaene-8,23-dione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wang, Y, Li, W. | | Deposit date: | 2021-02-16 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | C3 ester side chain plays a pivotal role in the antitumor activity of Maytansinoids.
Biochem.Biophys.Res.Commun., 566, 2021
|
|
3U1M
 
 | | Structure of the mRNA splicing complex component Cwc2 | | Descriptor: | Pre-mRNA-splicing factor CWC2, ZINC ION | | Authors: | Lu, P, Lu, G, Yan, C, Wang, L, Li, W, Yin, P. | | Deposit date: | 2011-09-30 | | Release date: | 2011-11-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the mRNA splicing complex component Cwc2: insights into RNA recognition
Biochem.J., 441, 2012
|
|
6K0R
 
 | | Ruvbl1-Ruvbl2 with truncated domain II in complex with phosphorylated Cordycepin | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Zhang, W, Chen, L, Li, W, Ju, D, Huang, N, Zhang, E. | | Deposit date: | 2019-05-07 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Chemical perturbations reveal that RUVBL2 regulates the circadian phase in mammals.
Sci Transl Med, 12, 2020
|
|
8J0K
 
 | | Crystal structure of human TFAP2A in complex with DNA | | Descriptor: | DNA (5'-D(*CP*TP*GP*CP*CP*TP*CP*GP*GP*GP*CP*AP*C)-3'), DNA (5'-D(*GP*TP*GP*CP*CP*CP*GP*AP*GP*GP*CP*AP*G)-3'), GLYCEROL, ... | | Authors: | Liu, K, Xiao, Y.Q, Li, W.F, Min, J.R. | | Deposit date: | 2023-04-11 | | Release date: | 2023-07-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for specific DNA sequence motif recognition by the TFAP2 transcription factors.
Nucleic Acids Res., 51, 2023
|
|
3U1L
 
 | | Structure of the mRNA splicing complex component Cwc2 | | Descriptor: | Pre-mRNA-splicing factor CWC2, ZINC ION | | Authors: | Lu, P, Lu, G, Yan, C, Wang, L, Li, W, Yin, P. | | Deposit date: | 2011-09-30 | | Release date: | 2011-11-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structure of the mRNA splicing complex component Cwc2: insights into RNA recognition
Biochem.J., 441, 2012
|
|
4LYC
 
 | | Cd ions within a lysoyzme single crystal | | Descriptor: | CADMIUM ION, Lysozyme C | | Authors: | Wei, H, House, S, Wu, J, Zhang, J, Wang, Z, He, Y, Gao, Y.-G, Robinson, H, Li, W, Zuo, J.-M, Robertson, I.M, Lu, Y. | | Deposit date: | 2013-07-30 | | Release date: | 2015-02-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Enhanced and tunable fluorescent quantum dots within a single crystal of protein
TO BE PUBLISHED
|
|
4LYB
 
 | | CdS within a lysoyzme single crystal | | Descriptor: | CADMIUM ION, Lysozyme C | | Authors: | Wei, H, House, S, Wu, J, Zhang, J, Wang, Z, He, Y, Gao, Y.-G, Robinson, H, Li, W, Zuo, J.-M, Robertson, I.M, Lu, Y. | | Deposit date: | 2013-07-30 | | Release date: | 2015-02-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Enhanced and tunable fluorescent quantum dots within a single crystal of protein
TO BE PUBLISHED
|
|
8TGI
 
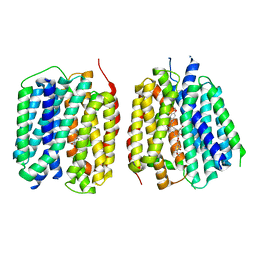 | | VMAT1 dimer with dopamine and reserpine | | Descriptor: | Chromaffin granule amine transporter, L-DOPAMINE, reserpine | | Authors: | Ye, J, Liu, B, Li, W. | | Deposit date: | 2023-07-12 | | Release date: | 2024-03-20 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into vesicular monoamine storage and drug interactions.
Nature, 629, 2024
|
|
8TGM
 
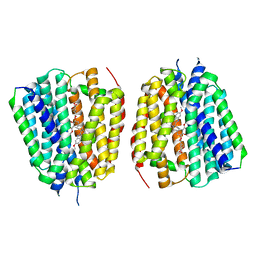 | | VMAT1 dimer with reserpine | | Descriptor: | VMAT1 dimer with reserpine, reserpine | | Authors: | Ye, J, Liu, B, Li, W. | | Deposit date: | 2023-07-12 | | Release date: | 2024-03-20 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into vesicular monoamine storage and drug interactions.
Nature, 629, 2024
|
|
8TGG
 
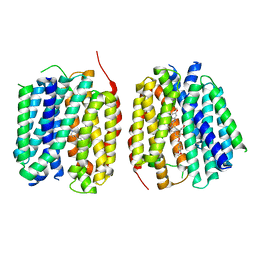 | | VMAT1 dimer with MPP+ and reserpine | | Descriptor: | 1-methyl-4-phenylpyridin-1-ium, Chromaffin granule amine transporter, reserpine | | Authors: | Ye, J, Liu, B, Li, W. | | Deposit date: | 2023-07-12 | | Release date: | 2024-03-20 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insights into vesicular monoamine storage and drug interactions.
Nature, 629, 2024
|
|
8TGH
 
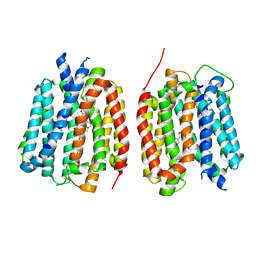 | | VMAT1 dimer with amphetamine and reserpine | | Descriptor: | (2S)-1-phenylpropan-2-amine, Chromaffin granule amine transporter, reserpine | | Authors: | Ye, J, Liu, B, Li, W. | | Deposit date: | 2023-07-12 | | Release date: | 2024-03-20 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into vesicular monoamine storage and drug interactions.
Nature, 629, 2024
|
|
8TGK
 
 | | VMAT1 dimer with histamine and reserpine | | Descriptor: | Chromaffin granule amine transporter, HISTAMINE, reserpine | | Authors: | Ye, J, Liu, B, Li, W. | | Deposit date: | 2023-07-12 | | Release date: | 2024-03-20 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural insights into vesicular monoamine storage and drug interactions.
Nature, 629, 2024
|
|
8TGJ
 
 | | VMAT1 dimer in unbound form and with reserpine | | Descriptor: | VMAT1 dimer in unbound form and with reserpine, reserpine | | Authors: | Ye, J, Liu, B, Li, W. | | Deposit date: | 2023-07-12 | | Release date: | 2024-03-20 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into vesicular monoamine storage and drug interactions.
Nature, 629, 2024
|
|
4OIC
 
 | | Crystal structrual of a soluble protein | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Bet v I allergen-like, CHLORIDE ION, ... | | Authors: | He, Y, Hao, Q, Li, W, Yan, C, Yan, N, Yin, P. | | Deposit date: | 2014-01-19 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Identification and characterization of ABA receptors in Oryza sativa
Plos One, 9, 2014
|
|
5Y2V
 
 | | Strcutrue of the full-length CcmR complexed with 2-OG from Synechocystis PCC6803 | | Descriptor: | 2-OXOGLUTARIC ACID, PHOSPHATE ION, Rubisco operon transcriptional regulator | | Authors: | Jiang, Y.L, Wang, X.P, Sun, H, Cheng, W, Han, S.J, Li, W.F, Chen, Y, Zhou, C.Z. | | Deposit date: | 2017-07-27 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Coordinating carbon and nitrogen metabolic signaling through the cyanobacterial global repressor NdhR.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5YL4
 
 | | CRYSTAL STRUCTURE OF T2R-TTL-8WR COMPLEX | | Descriptor: | (3Z,6Z)-3-[(4-tert-butyl-1H-imidazol-5-yl)methylidene]-6-[[3-(phenylcarbonyl)phenyl]methylidene]piperazine-2,5-dione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Fu, Z.Y, Li, W.B, Chu, Y.Y, Hou, Y.W, Ji, C.P. | | Deposit date: | 2017-10-17 | | Release date: | 2017-11-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Design, synthesis and biological activity evaluation of plinabulin derivatives based on co-crystal structure
To Be Published
|
|
