7F1E
 
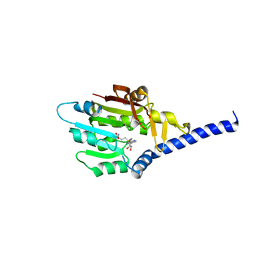 | | Structure of METTL6 bound with SAM | | Descriptor: | S-ADENOSYLMETHIONINE, tRNA N(3)-methylcytidine methyltransferase METTL6 | | Authors: | Li, S, Liao, S, Xu, C. | | Deposit date: | 2021-06-09 | | Release date: | 2022-01-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.589 Å) | | Cite: | Structural basis for METTL6-mediated m3C RNA methylation.
Biochem.Biophys.Res.Commun., 589, 2021
|
|
7UXU
 
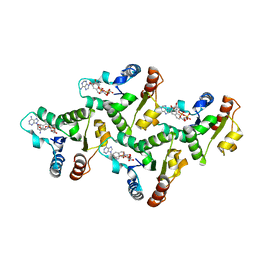 | | CryoEM structure of the TIR domain from AbTir in complex with 3AD | | Descriptor: | Molecular chaperone Tir, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-(8-azanylisoquinolin-2-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Li, S, Nanson, J.D, Manik, M.K, Gu, W, Landsberg, M.J, Ve, T, Kobe, B. | | Deposit date: | 2022-05-06 | | Release date: | 2022-09-07 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Cyclic ADP ribose isomers: Production, chemical structures, and immune signaling.
Science, 377, 2022
|
|
1SLL
 
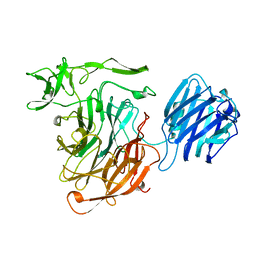 | | SIALIDASE L FROM LEECH MACROBDELLA DECORA | | Descriptor: | SIALIDASE L | | Authors: | Luo, Y, Li, S.C, Chou, M.Y, Li, Y.T, Luo, M. | | Deposit date: | 1997-10-14 | | Release date: | 1998-12-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of an intramolecular trans-sialidase with a NeuAc alpha2-->3Gal specificity.
Structure, 6, 1998
|
|
1SLI
 
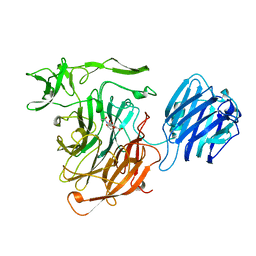 | | LEECH INTRAMOLECULAR TRANS-SIALIDASE COMPLEXED WITH DANA | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, INTRAMOLECULAR TRANS-SIALIDASE | | Authors: | Luo, Y, Li, S.C, Chou, M.Y, Li, Y.T, Luo, M. | | Deposit date: | 1998-03-28 | | Release date: | 1999-04-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of an intramolecular trans-sialidase with a NeuAc alpha2-->3Gal specificity.
Structure, 6, 1998
|
|
3UIU
 
 | | Crystal structure of Apo-PKR kinase domain | | Descriptor: | Interferon-induced, double-stranded RNA-activated protein kinase | | Authors: | Li, F, Li, S, Yang, X, Shen, Y, Zhang, T. | | Deposit date: | 2011-11-06 | | Release date: | 2012-11-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.903 Å) | | Cite: | Crystal structure of Apo-PKR kinase domain
TO BE PUBLISHED
|
|
5GZN
 
 | | Structure of neutralizing antibody bound to Zika envelope protein | | Descriptor: | Antibody Heavy chain, Antibody light chain, Genome polyprotein | | Authors: | Wang, Q, Yang, H, Liu, X, Dai, L, Ma, T, Qi, J, Wong, G, Peng, R, Liu, S, Li, J, Li, S, Song, J, Liu, J, He, J, Yuan, H, Xiong, Y, Liao, Y, Li, J, Yang, J, Tong, Z, Griffin, B, Bi, Y, Liang, M, Xu, X, Cheng, G, Wang, P, Qiu, X, Kobinger, G, Shi, Y, Yan, J, Gao, G.F. | | Deposit date: | 2016-09-29 | | Release date: | 2016-12-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular determinants of human neutralizing antibodies isolated from a patient infected with Zika virus
Sci Transl Med, 8, 2016
|
|
5HDE
 
 | | Crystal Structure of PTPN12 Catalytic Domain | | Descriptor: | PHOSPHATE ION, Tyrosine-protein phosphatase non-receptor type 12 | | Authors: | Dong, H, Li, S, Shi, J. | | Deposit date: | 2016-01-05 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal Structure of PTPN12 Catalytic Domain
To Be Published
|
|
2C6H
 
 | | Crystal structure of YC-17-bound cytochrome P450 PikC (CYP107L1) | | Descriptor: | 4-{[4-(DIMETHYLAMINO)-3-HYDROXY-6-METHYLTETRAHYDRO-2H-PYRAN-2-YL]OXY}-12-ETHYL-3,5,7,11-TETRAMETHYLOXACYCLODODEC-9-ENE-2,8-DIONE, CYTOCHROME P450 MONOOXYGENASE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Sherman, D.H, Li, S, Yermalitskaya, L.V, Kim, Y, Smith, J.A, Waterman, M.R, Podust, L.M. | | Deposit date: | 2005-11-09 | | Release date: | 2006-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Structural Basis for Substrate Anchoring, Active Site Selectivity, and Product Formation by P450 Pikc from Streptomyces Venezuelae.
J.Biol.Chem., 281, 2006
|
|
2BVJ
 
 | | Ligand-free structure of cytochrome P450 PikC (CYP107L1) | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, BETA-MERCAPTOETHANOL, CYTOCHROME P450 MONOOXYGENASE, ... | | Authors: | Sherman, D.H, Li, S, Yermalitskaya, L.V, Kim, Y, Smith, J.A, Waterman, M.R, Podust, L.M. | | Deposit date: | 2005-06-28 | | Release date: | 2006-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structural Basis for Substrate Anchoring, Active Site Selectivity, and Product Formation by P450 Pikc from Streptomyces Venezuelae.
J.Biol.Chem., 281, 2006
|
|
5GY3
 
 | |
4G8R
 
 | | Crystal Structure of a novel small molecule inactivator bound to plasminogen activator inhibitor-1 | | Descriptor: | (2S)-3-({[3-(trifluoromethyl)phenoxy]carbonyl}amino)propane-1,2-diyl bis(3,4,5-trihydroxybenzoate), Plasminogen activator inhibitor-1, SULFATE ION | | Authors: | Stuckey, J.A, Lawrence, D.A, Li, S.-H. | | Deposit date: | 2012-07-23 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Mechanistic characterization and crystal structure of a small molecule inactivator bound to plasminogen activator inhibitor-1.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4G8O
 
 | | Crystal Structure of a novel small molecule inactivator bound to plasminogen activator inhibitor-1 | | Descriptor: | (2S)-3-({[3-(trifluoromethyl)phenoxy]carbonyl}amino)propane-1,2-diyl bis(3,4,5-trihydroxybenzoate), 1,2-ETHANEDIOL, Plasminogen activator inhibitor 1, ... | | Authors: | Stuckey, J.A, Lawrence, D.A, Li, S.-H. | | Deposit date: | 2012-07-23 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Mechanistic characterization and crystal structure of a small molecule inactivator bound to plasminogen activator inhibitor-1.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4ORC
 
 | | Crystal structure of mammalian calcineurin | | Descriptor: | CALCIUM ION, Calcineurin subunit B type 1, FE (III) ION, ... | | Authors: | Ma, L, Li, S.J, Wang, J, Wu, J.W, Wang, Z.X. | | Deposit date: | 2014-02-11 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cooperative autoinhibition and multi-level activation mechanisms of calcineurin
To be Published
|
|
6VQV
 
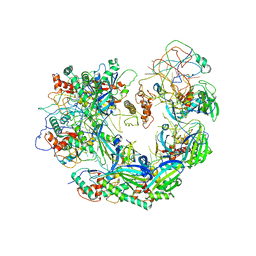 | | Type I-F CRISPR-Csy complex with its inhibitor AcrF9 | | Descriptor: | AcrF9, CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, ... | | Authors: | Zhang, K, Li, S, Pintilie, G, Zhu, Y, Huang, Z, Chiu, W. | | Deposit date: | 2020-02-06 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Inhibition mechanisms of AcrF9, AcrF8, and AcrF6 against type I-F CRISPR-Cas complex revealed by cryo-EM.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VQX
 
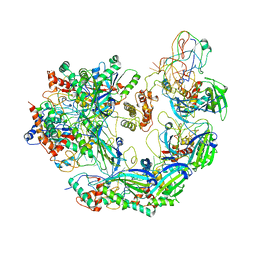 | | Type I-F CRISPR-Csy complex with its inhibitor AcrF6 | | Descriptor: | AcrF6, CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, ... | | Authors: | Zhang, K, Li, S, Pintilie, G, Zhu, Y, Huang, Z, Chiu, W. | | Deposit date: | 2020-02-06 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Inhibition mechanisms of AcrF9, AcrF8, and AcrF6 against type I-F CRISPR-Cas complex revealed by cryo-EM.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VQW
 
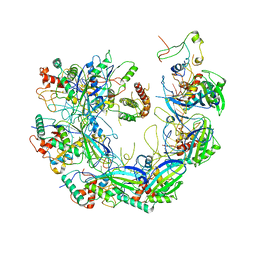 | | Type I-F CRISPR-Csy complex with its inhibitor AcrF8 | | Descriptor: | AcrF8, CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, ... | | Authors: | Zhang, K, Li, S, Pintilie, G, Zhu, Y, Huang, Z, Chiu, W. | | Deposit date: | 2020-02-06 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Inhibition mechanisms of AcrF9, AcrF8, and AcrF6 against type I-F CRISPR-Cas complex revealed by cryo-EM.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7YH8
 
 | | Crystal structure of a heterochiral protein complex | | Descriptor: | D-Pep-1, L-19437 | | Authors: | Liang, M, Li, S, Wang, T, Liu, L, Lu, P. | | Deposit date: | 2022-07-13 | | Release date: | 2023-07-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Accurate de novo design of heterochiral protein-protein interactions
Cell Res., 2024
|
|
9LAE
 
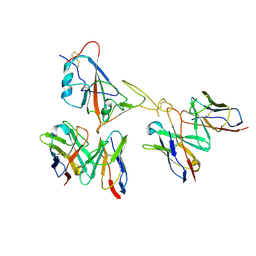 | | Locally refined region of SARS-CoV-2 spike in complex with antibodies 9G11 and 3E2. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 3E2, Heavy chain of 9G11, ... | | Authors: | Jiang, Y, Sun, H, Zheng, Q, Li, S. | | Deposit date: | 2025-01-02 | | Release date: | 2025-02-12 | | Last modified: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structural insight into broadening SARS-CoV-2 neutralization by an antibody cocktail harbouring both NTD and RBD potent antibodies.
Emerg Microbes Infect, 13, 2024
|
|
1SCZ
 
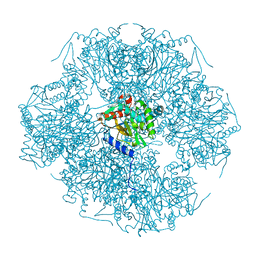 | | Improved structural model for the catalytic domain of E.coli dihydrolipoamide succinyltransferase | | Descriptor: | Dihydrolipoamide Succinyltransferase | | Authors: | Schormann, N, Symersky, J, Carson, M, Luo, M, Tsao, J, Johnson, D, Huang, W.-Y, Pruett, P, Lin, G, Li, S, Qiu, S, Arabashi, A, Bunzel, B, Luo, D, Nagy, L, Gray, R, Luan, C.-H, Zhang, Z, Lu, S, DeLucas, L. | | Deposit date: | 2004-02-12 | | Release date: | 2004-03-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Improved structural model for the catalytic domain of E.coli dihydrolipoamide succinyltransferase
To be Published
|
|
1SPX
 
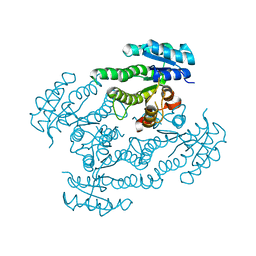 | | Crystal Structure of Glucose Dehydrogenase of Caenorhabditis Elegans in the Apo-Form | | Descriptor: | short-chain reductase family member (5L265) | | Authors: | Schormann, N, Zhou, J, McCombs, D, Bray, T, Symersky, J, Huang, W.-Y, Luan, C.-H, Gray, R, Luo, D, Arabashi, A, Bunzel, B, Nagy, L, Lu, S, Li, S, Lin, G, Zhang, Y, Qiu, S, Tsao, J, Luo, M, Carson, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-03-17 | | Release date: | 2004-03-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Glucose Dehydrogenase of Caenorhabditis Elegans in the Apo-Form: A Member of the SDR-Family
To be Published
|
|
7Y4W
 
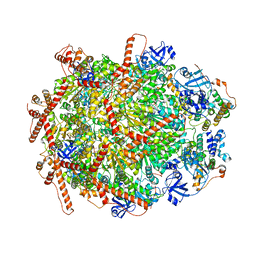 | | The cryo-EM structure of human ERAD retro-translocation complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Derlin-1, Transitional endoplasmic reticulum ATPase | | Authors: | Cao, Y, Rao, B, Wang, Q, Yao, D, Xia, Y, Li, W, Li, S, Shen, Y. | | Deposit date: | 2022-06-16 | | Release date: | 2023-10-18 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | The cryo-EM structure of the human ERAD retrotranslocation complex.
Sci Adv, 9, 2023
|
|
7YC8
 
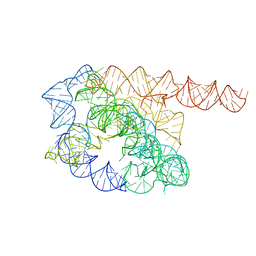 | | Cryo-EM structure of Tetrahymena ribozyme conformation 1 undergoing the first-step self-splicing | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, RNA (388-MER) | | Authors: | Zhang, X, Li, S, Pintilie, G, Palo, M.Z, Zhang, K. | | Deposit date: | 2022-07-01 | | Release date: | 2023-07-05 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (4.14 Å) | | Cite: | Snapshots of the first-step self-splicing of Tetrahymena ribozyme revealed by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
7YCI
 
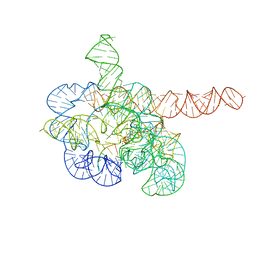 | | Cryo-EM structure of Tetrahymena ribozyme conformation 4 undergoing the first-step self-splicing | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, RNA (389-MER), ... | | Authors: | Zhang, X, Li, S, Pintilie, G, Palo, M.Z, Zhang, K. | | Deposit date: | 2022-07-01 | | Release date: | 2023-07-05 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Snapshots of the first-step self-splicing of Tetrahymena ribozyme revealed by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
7YCG
 
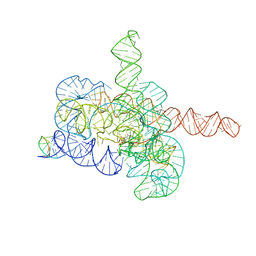 | | Cryo-EM structure of Tetrahymena ribozyme conformation 2 undergoing the first-step self-splicing | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, RNA (393-MER), ... | | Authors: | Zhang, X, Li, S, Pintilie, G, Palo, M.Z, Zhang, K. | | Deposit date: | 2022-07-01 | | Release date: | 2023-07-05 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Snapshots of the first-step self-splicing of Tetrahymena ribozyme revealed by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
7YCH
 
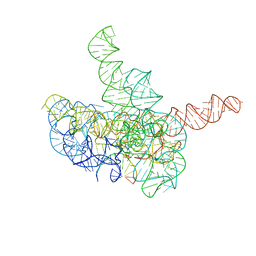 | | Cryo-EM structure of Tetrahymena ribozyme conformation 3 undergoing the first-step self-splicing | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, RNA (393-MER), ... | | Authors: | Zhang, X, Li, S, Pintilie, G, Palo, M.Z, Zhang, K. | | Deposit date: | 2022-07-01 | | Release date: | 2023-07-05 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Snapshots of the first-step self-splicing of Tetrahymena ribozyme revealed by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
