4LS1
 
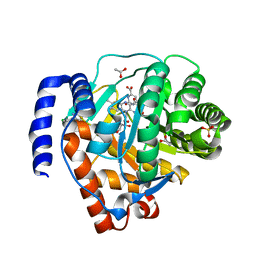 | | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with DH03A312 | | Descriptor: | 2-[(E)-{2-[4-(2-chlorophenyl)-1,3-thiazol-2-yl]hydrazinylidene}methyl]benzoic acid, ACETATE ION, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Zhu, L, Li, H, Ren, X, Zhu, J. | | Deposit date: | 2013-07-21 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with DH03A312
To be Published
|
|
4LWA
 
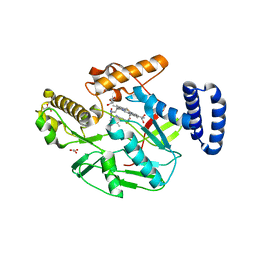 | | Structure of Bacillus subtilis nitric oxide synthase in complex with ((2S, 3S)-1,3-bis((6-(2,5-dimethyl-1H-pyrrol-1-yl)-4-methylpyridin-2-yl)methoxy)-2-aminobutane | | Descriptor: | 6,6'-{[(2S,3S)-2-aminobutane-1,3-diyl]bis(oxymethanediyl)}bis(4-methylpyridin-2-amine), CHLORIDE ION, GLYCEROL, ... | | Authors: | Holden, J.K, Li, H, Poulos, T.P. | | Deposit date: | 2013-07-26 | | Release date: | 2013-10-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural and biological studies on bacterial nitric oxide synthase inhibitors.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4MOZ
 
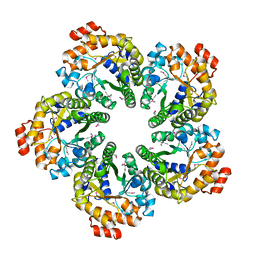 | |
7X8F
 
 | |
8D2N
 
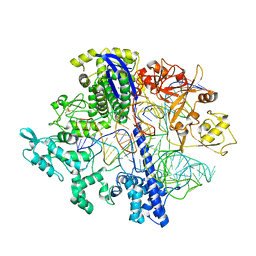 | | Structure of Acidothermus cellulolyticus Cas9 ternary complex (Pre-cleavage) | | Descriptor: | CRISPR-associated endonuclease, Csn1 family, DNA non-target strand (5'-D(P*TP*AP*CP*AP*CP*CP*AP*AP*GP*CP*T)-3'), ... | | Authors: | Rai, J, Das, A, Li, H. | | Deposit date: | 2022-05-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Coupled catalytic states and the role of metal coordination in Cas9.
Nat Catal, 6, 2023
|
|
6LPF
 
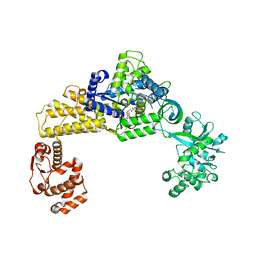 | | The crystal structure of human cytoplasmic LRS | | Descriptor: | 2'-(L-NORVALYL)AMINO-2'-DEOXYADENOSINE, 5'-O-(L-leucylsulfamoyl)adenosine, GLYCEROL, ... | | Authors: | Liu, R.J, Long, T, Li, H, Li, J, Zhao, J.H, Lin, J.Z, Palencia, A, Wang, M.Z, Cusack, S, Wang, E.D. | | Deposit date: | 2020-01-10 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Molecular basis of the multifaceted functions of human leucyl-tRNA synthetase in protein synthesis and beyond.
Nucleic Acids Res., 48, 2020
|
|
9ASI
 
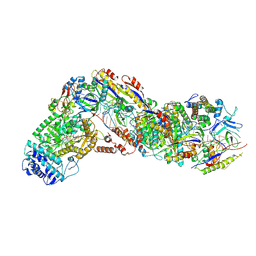 | | Cryo-EM structure of the active Lactococcus lactis Csm bound to target in pre-cleavage stage | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, CRISPR RNA, ... | | Authors: | Wang, B, Goswami, H.N, Li, H. | | Deposit date: | 2024-02-25 | | Release date: | 2024-08-14 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Molecular basis for cA6 synthesis by a type III-A CRISPR-Cas enzyme and its conversion to cA4 production.
Nucleic Acids Res., 52, 2024
|
|
9ASH
 
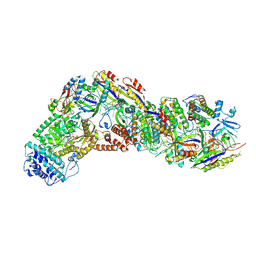 | | Cryo-EM structure of the active Lactococcus lactis Csm bound to target in post-cleavage stage | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CRISPR RNA, CRISPR system Cms endoribonuclease Csm3, ... | | Authors: | Wang, B, Goswami, H.N, Li, H. | | Deposit date: | 2024-02-25 | | Release date: | 2024-08-14 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Molecular basis for cA6 synthesis by a type III-A CRISPR-Cas enzyme and its conversion to cA4 production.
Nucleic Acids Res., 52, 2024
|
|
4MZH
 
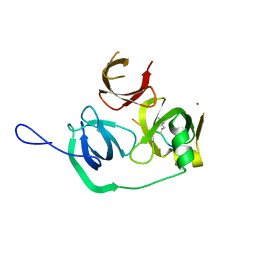 | |
8YAG
 
 | | Cryo-electron microscopic structure of an amide hydrolase from Pseudoxanthomonas wuyuanensis | | Descriptor: | Imidazolonepropionase, ZINC ION | | Authors: | Dai, L.H, Xu, Y.H, Hu, Y.M, Niu, D, Yang, X.C, Shen, P.P, Li, X, Xie, Z.Z, Li, H, Guo, R.-T, Chen, C.-C. | | Deposit date: | 2024-02-09 | | Release date: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Functional characterization and structural basis of an efficient ochratoxin A-degrading amidohydrolase.
Int.J.Biol.Macromol., 278, 2024
|
|
6U0M
 
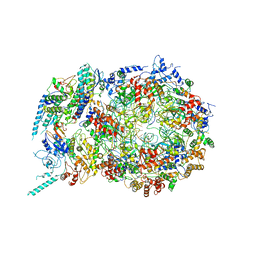 | | Structure of the S. cerevisiae replicative helicase CMG in complex with a forked DNA | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 45, DNA (15-MER), ... | | Authors: | Yuan, Z, Georgescu, R, Bai, L, Zhang, D, O'Donnell, M, Li, H. | | Deposit date: | 2019-08-14 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | DNA unwinding mechanism of a eukaryotic replicative CMG helicase.
Nat Commun, 11, 2020
|
|
8E1A
 
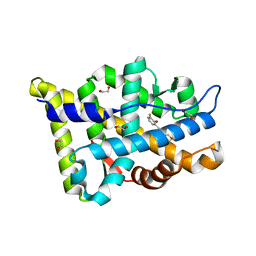 | | Structure-based study to overcome cross-reactivity of novel androgen receptor inhibitors | | Descriptor: | 1,2-ETHANEDIOL, 4-[4-(3-fluoro-2-methoxyphenyl)-1,3-thiazol-2-yl]morpholine, Androgen receptor | | Authors: | Lallous, N, Li, H, Radaeva, M, Dalal, K, Leblanc, E, Ban, F, Ciesielski, F, Chow, B, Morin, M, Singh, K, Rennie, P.S, Cherkasov, A. | | Deposit date: | 2022-08-10 | | Release date: | 2022-09-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure-Based Study to Overcome Cross-Reactivity of Novel Androgen Receptor Inhibitors.
Cells, 11, 2022
|
|
8K5R
 
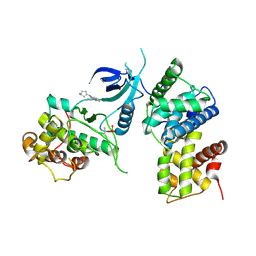 | | CDK9/cyclin T1 in complex with KB-0742 | | Descriptor: | (1S,3S)-N3-(5-pentan-3-ylpyrazolo[1,5-a]pyrimidin-7-yl)cyclopentane-1,3-diamine, Cyclin-T1, Cyclin-dependent kinase 9 | | Authors: | Zhou, M, Li, H, Gao, H, Trotter, B.W, Freeman, D. | | Deposit date: | 2023-07-24 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.751 Å) | | Cite: | Discovery of KB-0742, a Potent, Selective, Orally Bioavailable Small Molecule Inhibitor of CDK9 for MYC-Dependent Cancers.
J.Med.Chem., 66, 2023
|
|
6LR3
 
 | | Structural and functional insights into macrophage migration inhibitory factor from Oncomelania hupensis, the intermediate host of Schistosoma japonicum | | Descriptor: | Macrophage migration inhibitory factor, SULFATE ION | | Authors: | Su, Z.M, Tian, X.Y, Li, H.J, Wei, Z.M, Chen, L.F, Ren, H.X, Peng, W.F, Tang, C.T. | | Deposit date: | 2020-01-15 | | Release date: | 2020-07-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural and functional insights into macrophage migration inhibitory factor from Oncomelania hupensis, the intermediate host of Schistosoma japonicum.
Biochem.J., 477, 2020
|
|
6LKV
 
 | | Structural and functional insights into macrophage migration inhibitory factor from Oncomelania hupensis, the intermediate host of Schistosoma japonicum | | Descriptor: | CHLORIDE ION, Macrophage migration inhibitory factor, SULFATE ION | | Authors: | Su, Z.M, Tian, X.Y, Li, H.J, Wei, Z.M, Chen, L.F, Ren, H.X, Peng, W.F, Tang, C.T. | | Deposit date: | 2019-12-20 | | Release date: | 2020-07-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional insights into macrophage migration inhibitory factor from Oncomelania hupensis, the intermediate host of Schistosoma japonicum.
Biochem.J., 477, 2020
|
|
6WEJ
 
 | | Structure of cGMP-unbound WT TAX-4 reconstituted in lipid nanodiscs | | Descriptor: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Cyclic nucleotide-gated cation channel, ... | | Authors: | Zheng, X, Fu, Z, Su, D, Zhang, Y, Li, M, Pan, Y, Li, H, Li, S, Grassucci, R.A, Ren, Z, Hu, Z, Li, X, Zhou, M, Li, G, Frank, J, Yang, J. | | Deposit date: | 2020-04-02 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-22 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Mechanism of ligand activation of a eukaryotic cyclic nucleotide-gated channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6WEK
 
 | | Structure of cGMP-bound WT TAX-4 reconstituted in lipid nanodiscs | | Descriptor: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CYCLIC GUANOSINE MONOPHOSPHATE, ... | | Authors: | Zheng, X, Fu, Z, Su, D, Zhang, Y, Li, M, Pan, Y, Li, H, Li, S, Grassucci, R.A, Ren, Z, Hu, Z, Li, X, Zhou, M, Li, G, Frank, J, Yang, J. | | Deposit date: | 2020-04-02 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-22 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Mechanism of ligand activation of a eukaryotic cyclic nucleotide-gated channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6WEL
 
 | | Structure of cGMP-unbound F403V/V407A mutant TAX-4 reconstituted in lipid nanodiscs | | Descriptor: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Cyclic nucleotide-gated cation channel, ... | | Authors: | Zheng, X, Fu, Z, Su, D, Zhang, Y, Li, M, Pan, Y, Li, H, Li, S, Grassucci, R.A, Ren, Z, Hu, Z, Li, X, Zhou, M, Li, G, Frank, J, Yang, J. | | Deposit date: | 2020-04-02 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Mechanism of ligand activation of a eukaryotic cyclic nucleotide-gated channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
5J9S
 
 | | ENL YEATS in complex with histone H3 acetylation at K27 | | Descriptor: | H3K27ac peptide from Histone H3.1, Protein ENL, SULFATE ION | | Authors: | Li, Y, Li, H. | | Deposit date: | 2016-04-11 | | Release date: | 2017-03-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | ENL links histone acetylation to oncogenic gene expression in acute myeloid leukaemia
Nature, 543, 2017
|
|
8NSE
 
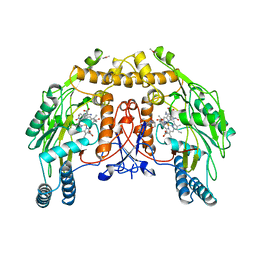 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE, NNA COMPLEX | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, CACODYLIC ACID, GLYCEROL, ... | | Authors: | Raman, C.S, Li, H, Martasek, P, Masters, B.S.S, Poulos, T.L. | | Deposit date: | 1999-01-14 | | Release date: | 2001-11-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of nitric oxide synthase bound to nitro indazole reveals a novel inactivation mechanism.
Biochemistry, 40, 2001
|
|
6LLX
 
 | |
9B8T
 
 | | Human polymerase epsilon bound to PCNA and DNA in the nucleotide bound state | | Descriptor: | DNA polymerase epsilon catalytic subunit, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Wang, F, He, Q, Li, H. | | Deposit date: | 2024-03-31 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structures of the human leading strand Pol epsilon-PCNA holoenzyme.
Nat Commun, 15, 2024
|
|
5J3J
 
 | | Crystal Structure of human DPP-IV in complex with HL1 | | Descriptor: | (2~{S},3~{R})-8,9-dimethoxy-3-[2,4,5-tris(fluoranyl)phenyl]-2,3-dihydro-1~{H}-benzo[f]chromen-2-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4, ... | | Authors: | Wu, F, Li, H, Zhao, Z, Zhu, L, Xu, H, Li, S. | | Deposit date: | 2016-03-31 | | Release date: | 2017-04-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structure of human DPP-IV in complex with HL1
To Be Published
|
|
7XT9
 
 | | Serotonin 4 (5-HT4) receptor-Gs complex | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Huang, S, Xu, P, Shen, D.D, Simon, I.A, Mao, C, Tan, Y, Zhang, H, Harpsoe, K, Li, H, Zhang, Y, You, C, Yu, X, Jiang, Y, Zhang, Y, Gloriam, D.E, Xu, H.E. | | Deposit date: | 2022-05-16 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | GPCRs steer G i and G s selectivity via TM5-TM6 switches as revealed by structures of serotonin receptors.
Mol.Cell, 82, 2022
|
|
7XT8
 
 | | Serotonin 4 (5-HT4) receptor-Gs-Nb35 complex | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Huang, S, Xu, P, Shen, D.D, Simon, I.A, Mao, C, Tan, Y, Zhang, H, Harpsoe, K, Li, H, Zhang, Y, You, C, Yu, X, Jiang, Y, Zhang, Y, Gloriam, D.E, Xu, H.E. | | Deposit date: | 2022-05-16 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | GPCRs steer G i and G s selectivity via TM5-TM6 switches as revealed by structures of serotonin receptors.
Mol.Cell, 82, 2022
|
|
