5KT2
 
 | | Teranry complex of human DNA polymerase iota(26-445) inserting dCMPNPP opposite template G in the presence of Mg2+ | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, DNA (5'-D(*CP*TP*GP*GP*GP*GP*TP*CP*CP*T)-3'), DNA (5'-D(P*AP*GP*GP*AP*CP*CP*C)-3'), ... | | Authors: | Choi, J.Y, Patra, A, Yeom, M, Lee, Y.S, Zhang, Q, Egli, M, Guengerich, F.P. | | Deposit date: | 2016-07-11 | | Release date: | 2016-08-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.488 Å) | | Cite: | Kinetic and Structural Impact of Metal Ions and Genetic Variations on Human DNA Polymerase iota.
J.Biol.Chem., 291, 2016
|
|
5KT7
 
 | | Teranry complex of human DNA polymerase iota(1-445) inserting dCMPNPP opposite template G in the presence of Mn2+ | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, DNA (5'-D(P*AP*GP*GP*AP*CP*CP*C)-3'), DNA (5'-D(P*TP*GP*GP*GP*GP*TP*CP*CP*T)-3'), ... | | Authors: | Choi, J.Y, Patra, A, Yeom, M, Lee, Y.S, Zhang, Q, Egli, M, Guengerich, F.P. | | Deposit date: | 2016-07-11 | | Release date: | 2016-08-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.151 Å) | | Cite: | Kinetic and Structural Impact of Metal Ions and Genetic Variations on Human DNA Polymerase iota.
J.Biol.Chem., 291, 2016
|
|
5KT6
 
 | | Teranry complex of human DNA polymerase iota(1-445) inserting dCMPNPP opposite template G in the presence of Mg2+ | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, DNA (5'-D(P*AP*GP*GP*AP*CP*CP*C)-3'), DNA (5'-D(P*TP*GP*GP*GP*GP*TP*CP*CP*T)-3'), ... | | Authors: | Choi, J.Y, Patra, A, Yeom, M, Lee, Y.S, Zhang, Q, Egli, M, Guengerich, F.P. | | Deposit date: | 2016-07-11 | | Release date: | 2016-08-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.54 Å) | | Cite: | Kinetic and Structural Impact of Metal Ions and Genetic Variations on Human DNA Polymerase iota.
J.Biol.Chem., 291, 2016
|
|
5KT3
 
 | | Teranry complex of human DNA polymerase iota(26-445) inserting dCMPNPP opposite template G in the presence of Mn2+ | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, DNA (5'-D(*CP*TP*GP*GP*GP*GP*TP*CP*CP*T)-3'), DNA (5'-D(P*AP*GP*GP*AP*CP*CP*C)-3'), ... | | Authors: | Choi, J.Y, Patra, A, Yeom, M, Lee, Y.S, Zhang, Q, Egli, M, Guengerich, F.P. | | Deposit date: | 2016-07-11 | | Release date: | 2016-08-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Kinetic and Structural Impact of Metal Ions and Genetic Variations on Human DNA Polymerase iota.
J.Biol.Chem., 291, 2016
|
|
5KT4
 
 | | Teranry complex of human DNA polymerase iota R96G inserting dCMPNPP opposite template G in the presence of Mg2+ | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, DNA (5'-D(*CP*TP*GP*GP*GP*GP*TP*CP*CP*T)-3'), DNA (5'-D(P*AP*GP*GP*AP*CP*CP*C)-3'), ... | | Authors: | Choi, J.Y, Patra, A, Yeom, M, Lee, Y.S, Zhang, Q, Egli, M, Guengerich, F.P. | | Deposit date: | 2016-07-11 | | Release date: | 2016-08-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Kinetic and Structural Impact of Metal Ions and Genetic Variations on Human DNA Polymerase iota.
J.Biol.Chem., 291, 2016
|
|
5KT5
 
 | | Teranry complex of human DNA polymerase iota R96G inserting dCMPNPP opposite template G in the presence of Mn2+ | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, DNA (5'-D(*CP*TP*GP*GP*GP*GP*TP*CP*CP*T)-3'), DNA (5'-D(P*AP*GP*GP*AP*CP*CP*C)-3'), ... | | Authors: | Choi, J.Y, Patra, A, Yeom, M, Lee, Y.S, Zhang, Q, Egli, M, Guengerich, F.P. | | Deposit date: | 2016-07-11 | | Release date: | 2016-08-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.798 Å) | | Cite: | Kinetic and Structural Impact of Metal Ions and Genetic Variations on Human DNA Polymerase iota.
J.Biol.Chem., 291, 2016
|
|
6PWV
 
 | | Cryo-EM structure of MLL1 core complex bound to the nucleosome | | Descriptor: | DNA (147-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Park, S.H, Ayoub, A, Lee, Y.T, Xu, J, Zhang, W, Zhang, B, Zhang, Y, Cianfrocco, M.A, Su, M, Dou, Y, Cho, U. | | Deposit date: | 2019-07-23 | | Release date: | 2019-12-18 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Cryo-EM structure of the human MLL1 core complex bound to the nucleosome.
Nat Commun, 10, 2019
|
|
2VW9
 
 | | Single stranded DNA binding protein complex from Helicobacter pylori | | Descriptor: | POLY-DT, SINGLE-STRANDED DNA BINDING PROTEIN | | Authors: | Chan, K.-W, Wang, C.-H, Lee, Y.-J, Sun, Y.-J. | | Deposit date: | 2008-06-18 | | Release date: | 2009-06-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Single-Stranded DNA-Binding Protein Complex from Helicobacter Pylori Suggests an Ssdna-Binding Surface.
J.Mol.Biol., 388, 2009
|
|
5IK1
 
 | |
6JB7
 
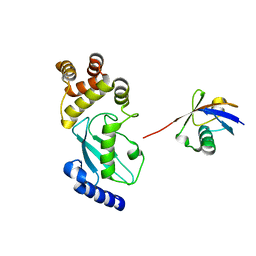 | | Crystal structure of Ub-conjugated Ube2K C92K&K97A mutant (isopeptide linkage), 2.1 A resolution | | Descriptor: | Ubiquitin, Ubiquitin-conjugating enzyme E2 K | | Authors: | Lee, J.-G, Youn, H.-S, Lee, Y, An, J.Y, Park, K.R, Kang, J.Y, Mun, S.A, Park, J, Park, T, Jin, M.W, Yang, J, Eom, S.H. | | Deposit date: | 2019-01-25 | | Release date: | 2019-03-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of Ub-conjugated Ube2K C92K&K97A mutant (isopeptide linkage), 2.1 A resolution
To Be Published
|
|
2AXN
 
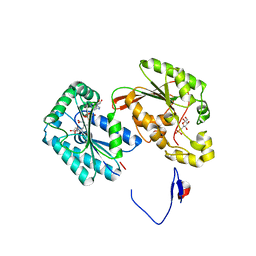 | | Crystal structure of the human inducible form 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 (6PF-2-K/Fru- 2,6-P2ASE brain/placenta-type isozyme) (iPFK-2) [Includes: 6- phosphofructo-2-kinase (EC 2.7.1.105); Fructose-2,6-bisphosphatase (EC 3.1.3.46)], ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Kim, S.G, Manes, N.P, El-Maghrabi, M.R, Lee, Y.H. | | Deposit date: | 2005-09-05 | | Release date: | 2005-12-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the hypoxia-inducible form of 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase (PFKFB3): a possible new target for cancer therapy.
J.Biol.Chem., 281, 2006
|
|
6PWW
 
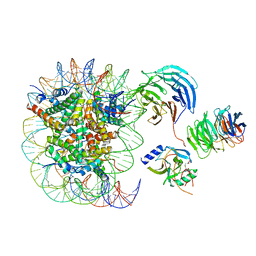 | | Cryo-EM structure of MLL1 in complex with RbBP5 and WDR5 bound to the nucleosome | | Descriptor: | DNA (146-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Park, S.H, Ayoub, A, Lee, Y.T, Xu, J, Zhang, W, Zhang, B, Zhang, Y, Cianfrocco, M.A, Su, M, Dou, Y, Cho, U. | | Deposit date: | 2019-07-23 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM structure of the human MLL1 core complex bound to the nucleosome.
Nat Commun, 10, 2019
|
|
6PWX
 
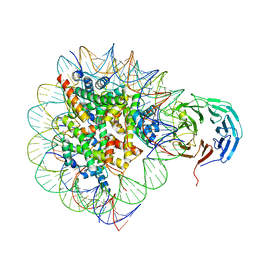 | | Cryo-EM structure of RbBP5 bound to the nucleosome | | Descriptor: | DNA (146-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Park, S.H, Ayoub, A, Lee, Y.T, Xu, J, Zhang, W, Zhang, B, Zhang, Y, Cianfrocco, M.A, Su, M, Dou, Y, Cho, U. | | Deposit date: | 2019-07-23 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structure of the human MLL1 core complex bound to the nucleosome.
Nat Commun, 10, 2019
|
|
8WG4
 
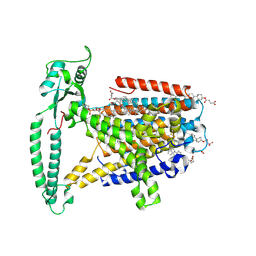 | | mouse TMEM63b in DDM-CHS micelle with YN9303-24 Fab | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, CHOLESTEROL HEMISUCCINATE, CSC1-like protein 2,Green fluorescent protein | | Authors: | Miyata, Y, Takahashi, K, Lee, Y, Sultan, C.S, Kuribayashi, R, Takahashi, M, Hata, K, Bamba, T, Izumi, Y, Liu, K, Uemura, T, Nomura, N, Iwata, S, Nagata, S, Nishizawa, T, Segawa, K. | | Deposit date: | 2023-09-20 | | Release date: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mechanosensitive channel TMEM63B functions as a plasma membrane lipid scramblase
To Be Published
|
|
8WG3
 
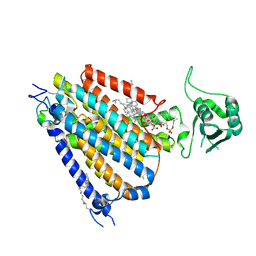 | | mouse TMEM63b in LMNG-CHS micelle | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, CHOLESTEROL HEMISUCCINATE, CSC1-like protein 2,Green fluorescent protein | | Authors: | Miyata, Y, Takahashi, K, Lee, Y, Sultan, C.S, Kuribayashi, R, Takahashi, M, Hata, K, Bamba, T, Izumi, Y, Liu, K, Uemura, T, Nomura, N, Iwata, S, Nagata, S, Nishizawa, T, Segawa, K. | | Deposit date: | 2023-09-20 | | Release date: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanosensitive channel TMEM63B functions as a plasma membrane lipid scramblase
To Be Published
|
|
1XVJ
 
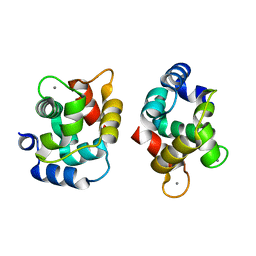 | | Crystal Structure Of Rat alpha-Parvalbumin D94S/G98E Mutant | | Descriptor: | CALCIUM ION, Parvalbumin alpha | | Authors: | Tanner, J.J, Agah, S, Lee, Y.H, Henzl, M.T. | | Deposit date: | 2004-10-28 | | Release date: | 2005-09-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the D94S/G98E Variant of Rat alpha-Parvalbumin. An Explanation for the Reduced Divalent Ion Affinity.
Biochemistry, 44, 2005
|
|
1Y93
 
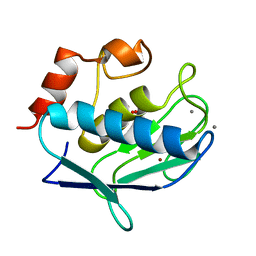 | | Crystal structure of the catalytic domain of human MMP12 complexed with acetohydroxamic acid at atomic resolution | | Descriptor: | ACETOHYDROXAMIC ACID, CALCIUM ION, Macrophage metalloelastase, ... | | Authors: | Bertini, I, Calderone, V, Cosenza, M, Fragai, M, Lee, Y.-M, Luchinat, C, Mangani, S, Terni, B, Turano, P. | | Deposit date: | 2004-12-14 | | Release date: | 2005-04-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Conformational variability of matrix metalloproteinases: Beyond a single 3D structure
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1YSM
 
 | | NMR Structure of N-terminal domain (Residues 1-77) of Siah-Interacting Protein. | | Descriptor: | Calcyclin-binding protein | | Authors: | Bhattacharya, S, Lee, Y.T, Michowski, W, Jastrzebska, B, Filipek, A, Kuznicki, J, Chazin, W.J. | | Deposit date: | 2005-02-08 | | Release date: | 2005-07-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The Modular Structure of SIP Facilitates Its Role in Stabilizing Multiprotein Assemblies.
Biochemistry, 44, 2005
|
|
4GAV
 
 | | Structure of the Ndi1 protein from Saccharomyces cerevisiae in complex with quinone | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Rotenone-insensitive NADH-ubiquinone oxidoreductase, UBIQUINONE-2 | | Authors: | Iwata, M, Lee, Y, Yamashita, T, Yagi, T, Iwata, S, Cameron, A.D, Maher, M.J. | | Deposit date: | 2012-07-25 | | Release date: | 2012-09-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of the yeast NADH dehydrogenase (Ndi1) reveals overlapping binding sites for water- and lipid-soluble substrates.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4G9K
 
 | | Structure of the Ndi1 protein from Saccharomyces cerevisiae | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Rotenone-insensitive NADH-ubiquinone oxidoreductase | | Authors: | Iwata, M, Lee, Y, Yamashita, T, Yagi, T, Iwata, S, Cameron, A.D, Maher, M.J. | | Deposit date: | 2012-07-24 | | Release date: | 2012-09-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of the yeast NADH dehydrogenase (Ndi1) reveals overlapping binding sites for water- and lipid-soluble substrates.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4GAP
 
 | | Structure of the Ndi1 protein from Saccharomyces cerevisiae in complex with NAD+ | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Rotenone-insensitive NADH-ubiquinone oxidoreductase | | Authors: | Iwata, M, Lee, Y, Yamashita, T, Yagi, T, Iwata, S, Cameron, A.D, Maher, M.J. | | Deposit date: | 2012-07-25 | | Release date: | 2012-09-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure of the yeast NADH dehydrogenase (Ndi1) reveals overlapping binding sites for water- and lipid-soluble substrates.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1RK9
 
 | | Solution Structure of Human alpha-Parvalbumin (Minimized Average Structure) | | Descriptor: | CALCIUM ION, Parvalbumin alpha | | Authors: | Baig, I, Bertini, I, Del Bianco, C, Gupta, Y.K, Lee, Y.-M, Luchinat, C, Quattrone, A, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-11-21 | | Release date: | 2004-06-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Paramagnetism-based refinement strategy for the solution structure of human alpha-parvalbumin
Biochemistry, 43, 2004
|
|
8PVA
 
 | | Structure of bacterial ribosome determined by cryoEM at 100 keV | | Descriptor: | 16S rRNA, 23S rRNA, 50S ribosomal protein L14, ... | | Authors: | McMullan, G, Naydenova, K, Mihaylov, D, Peet, M.J, Wilson, H, Yamashita, K, Dickerson, J.L, Chen, S, Cannone, G, Lee, Y, Hutchings, K.A, Gittins, O, Sobhy, M, Wells, T, El-Gomati, M.M, Dalby, J, Meffert, M, Schulze-Briese, C, Henderson, R, Russo, C.J. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-29 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structure determination by cryoEM at 100 keV.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
2O5K
 
 | | Crystal Structure of GSK3beta in complex with a benzoimidazol inhibitor | | Descriptor: | 2-(2,4-DICHLORO-PHENYL)-7-HYDROXY-1H-BENZOIMIDAZOLE-4-CARBOXYLIC ACID [2-(4-METHANESULFONYLAMINO-PHENYL)-ETHYL]-AMIDE, Glycogen synthase kinase-3 beta | | Authors: | Shin, D, Lee, S.C, Heo, Y.S, Cho, Y.S, Kim, Y.E, Hyun, Y.L, Cho, J.M, Lee, Y.S, Ro, S. | | Deposit date: | 2006-12-06 | | Release date: | 2007-10-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Design and synthesis of 7-hydroxy-1H-benzoimidazole derivatives as novel inhibitors of glycogen synthase kinase-3beta
Bioorg.Med.Chem.Lett., 17, 2007
|
|
1RJV
 
 | | Solution Structure of Human alpha-Parvalbumin refined with a paramagnetism-based strategy | | Descriptor: | CALCIUM ION, Parvalbumin alpha | | Authors: | Baig, I, Bertini, I, Del Bianco, C, Gupta, Y.K, Lee, Y.M, Luchinat, C, Quattrone, A, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-11-20 | | Release date: | 2004-05-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Paramagnetism-Based Refinement Strategy for the Solution Structure of Human alpha-Parvalbumin.
Biochemistry, 43, 2004
|
|
