8K4M
 
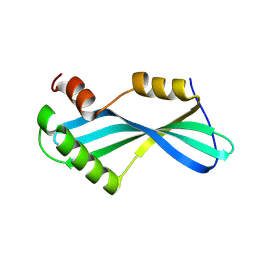 | | Anti CRISPR protein, AcrIIA13b | | Descriptor: | Anti CRISPR protein | | Authors: | Lee, S.Y, Park, H.H. | | Deposit date: | 2023-07-19 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structure of AcrIIA13b at 1.53 Angstroms resolution.
To Be Published
|
|
1F4V
 
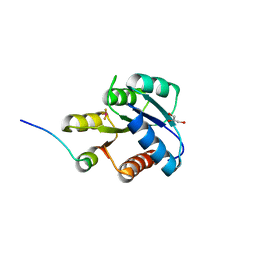 | | CRYSTAL STRUCTURE OF ACTIVATED CHEY BOUND TO THE N-TERMINUS OF FLIM | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, CHEMOTAXIS CHEY PROTEIN, FLAGELLAR MOTOR SWITCH PROTEIN, ... | | Authors: | Lee, S.Y, Cho, H.S, Pelton, J.G, Yan, D, Henderson, R.K, King, D, Huang, L.S, Kustu, S, Berry, E.A, Wemmer, D.E. | | Deposit date: | 2000-06-10 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal structure of an activated response regulator bound to its target.
Nat.Struct.Biol., 8, 2001
|
|
2A0L
 
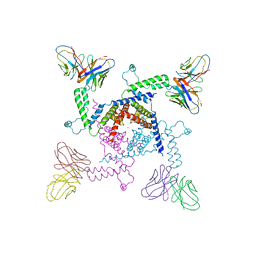 | | Crystal structure of KvAP-33H1 Fv complex | | Descriptor: | 33H1 Fv fragment, POTASSIUM ION, Voltage-gated potassium channel | | Authors: | Lee, S.Y, Lee, A, Chen, J, Mackinnon, R. | | Deposit date: | 2005-06-16 | | Release date: | 2005-11-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structure of the KvAP voltage-dependent K+ channel and its dependence on the lipid membrane.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1FQW
 
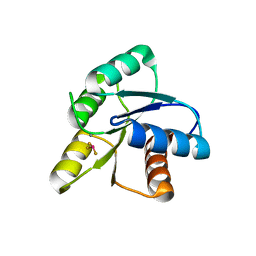 | | CRYSTAL STRUCTURE OF ACTIVATED CHEY | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, CHEMOTAXIS CHEY PROTEIN, MANGANESE (II) ION | | Authors: | Lee, S.Y, Cho, H.S, Pelton, J.G, Yan, D, Berry, E.A, Wemmer, D.E. | | Deposit date: | 2000-09-07 | | Release date: | 2001-07-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystal structure of activated CheY. Comparison with other activated receiver domains.
J.Biol.Chem., 276, 2001
|
|
1NY6
 
 | | Crystal structure of sigm54 activator (AAA+ ATPase) in the active state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, transcriptional regulator (NtrC family) | | Authors: | Lee, S.Y, de la Torre, A, Kustu, S, Nixon, B.T, Wemmer, D.E. | | Deposit date: | 2003-02-11 | | Release date: | 2003-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Regulation of the transcriptional activator NtrC1: structural studies of the regulatory and AAA+ ATPase domains
Genes Dev., 17, 2003
|
|
1NY5
 
 | | Crystal structure of sigm54 activator (AAA+ ATPase) in the inactive state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Lee, S.Y, de la Torre, A, Kustu, S, Nixon, B.T, Wemmer, D.E. | | Deposit date: | 2003-02-11 | | Release date: | 2003-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Regulation of the transcriptional activator NtrC1: structural studies of the regulatory and AAA+ ATPase domains
Genes Dev., 17, 2003
|
|
7V6E
 
 | | DREP3 | | Descriptor: | DNAation factor-related protein 3, isoform A | | Authors: | Lee, S.Y, Park, H.H. | | Deposit date: | 2021-08-20 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Helical filament structure of the DREP3 CIDE domain reveals a unified mechanism of CIDE-domain assembly.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5CKR
 
 | | Crystal Structure of MraY in complex with Muraymycin D2 | | Descriptor: | Muraymycin D2, Phospho-N-acetylmuramoyl-pentapeptide-transferase | | Authors: | Lee, S.Y, Chung, B.C, Mashalidis, E.H, Tanino, T, Kim, M, Hong, J, Ichikawa, S. | | Deposit date: | 2015-07-15 | | Release date: | 2016-03-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural insights into inhibition of lipid I production in bacterial cell wall synthesis.
Nature, 533, 2016
|
|
4J72
 
 | | Crystal Structure of polyprenyl-phosphate N-acetyl hexosamine 1-phosphate transferase | | Descriptor: | MAGNESIUM ION, NICKEL (II) ION, Phospho-N-acetylmuramoyl-pentapeptide-transferase | | Authors: | Lee, S.Y, Chung, B.C, Gillespie, R.A, Kwon, D.Y, Guan, Z, Zhou, P, Hong, J. | | Deposit date: | 2013-02-12 | | Release date: | 2013-09-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of MraY, an essential membrane enzyme for bacterial cell wall synthesis.
Science, 341, 2013
|
|
7VI8
 
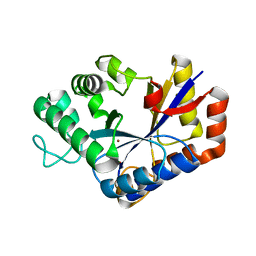 | | Crystal structure of ChbG | | Descriptor: | ACETATE ION, Chitooligosaccharide deacetylase, ZINC ION | | Authors: | Lee, S.Y, Park, H.H. | | Deposit date: | 2021-09-26 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structure of ChbG from Klebsiella pneumoniae reveals the molecular basis of diacetylchitobiose deacetylation.
Commun Biol, 5, 2022
|
|
7XI5
 
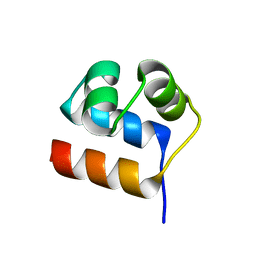 | | Anti-CRISPR-associated Aca10 | | Descriptor: | Transcriptional regulator | | Authors: | Lee, S.Y, Park, H.H. | | Deposit date: | 2022-04-12 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Molecular basis of anti-CRISPR operon repression by Aca10.
Nucleic Acids Res., 50, 2022
|
|
7CHQ
 
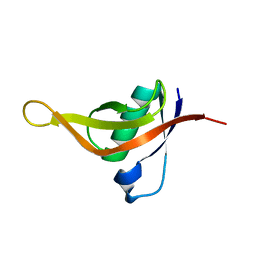 | | AcrIE2 | | Descriptor: | anti-CRISPR AcrIE2 | | Authors: | Lee, S.Y, Park, H.H. | | Deposit date: | 2020-07-06 | | Release date: | 2021-05-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | A 1.3 angstrom high-resolution crystal structure of an anti-CRISPR protein, AcrI E2.
Biochem.Biophys.Res.Commun., 533, 2020
|
|
7EZY
 
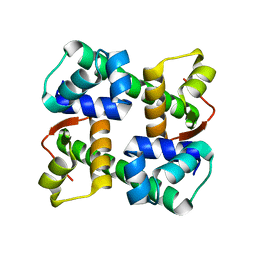 | | anti-CRISPR-associated Aca2 | | Descriptor: | anti-CRISPR-associated Aca2 | | Authors: | Lee, S.Y, Park, H.H. | | Deposit date: | 2021-06-02 | | Release date: | 2022-01-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Molecular basis of transcriptional repression ofanti-CRISPR by anti-CRISPR-associated 2
Acta Crystallogr.,Sect.D, 78, 2022
|
|
4KM5
 
 | | X-ray crystal structure of human cyclic GMP-AMP synthase (cGAS) | | Descriptor: | Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Kranzusch, P.J, Lee, A.S.Y, Berger, J.M, Doudna, J.A. | | Deposit date: | 2013-05-08 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Structure of Human cGAS Reveals a Conserved Family of Second-Messenger Enzymes in Innate Immunity.
Cell Rep, 3, 2013
|
|
4TY0
 
 | | Crystal structure of Vibrio cholerae DncV cyclic AMP-GMP synthase in complex with linear intermediate 5' pppA(3',5')pG | | Descriptor: | ACETATE ION, Cyclic AMP-GMP synthase, MAGNESIUM ION, ... | | Authors: | Kranzusch, P.J, Lee, A.S.Y, Wilson, S.C, Solovykh, M.S, Vance, R.E, Berger, J.M, Doudna, J.A. | | Deposit date: | 2014-07-07 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Guided Reprogramming of Human cGAS Dinucleotide Linkage Specificity.
Cell, 158, 2014
|
|
4TXZ
 
 | | Crystal structure of Vibrio cholerae DncV cyclic AMP-GMP synthase in complex with nonhydrolyzable GTP | | Descriptor: | Cyclic AMP-GMP synthase, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | Authors: | Kranzusch, P.J, Lee, A.S.Y, Wilson, S.C, Solovykh, M.S, Vance, R.E, Berger, J.M, Doudna, J.A. | | Deposit date: | 2014-07-07 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Guided Reprogramming of Human cGAS Dinucleotide Linkage Specificity.
Cell, 158, 2014
|
|
1DJM
 
 | | SOLUTION STRUCTURE OF BEF3-ACTIVATED CHEY FROM ESCHERICHIA COLI | | Descriptor: | CHEMOTAXIS PROTEIN Y | | Authors: | Cho, H.S, Lee, S.Y, Yan, D, Pan, X, Parkinson, J.S, Kustu, S, Wemmer, D.E, Pelton, J.G. | | Deposit date: | 1999-12-03 | | Release date: | 2000-04-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of activated CheY.
J.Mol.Biol., 297, 2000
|
|
5GPG
 
 | | Co-crystal structure of the FK506 binding domain of human FKBP25, Rapamycin and the FRB domain of human mTOR | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP3, RAPAMYCIN IMMUNOSUPPRESSANT DRUG, Serine/threonine-protein kinase mTOR | | Authors: | Lee, H.B, Lee, S.Y, Rhee, H.W, Lee, C.W. | | Deposit date: | 2016-08-02 | | Release date: | 2016-10-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Proximity-Directed Labeling Reveals a New Rapamycin-Induced Heterodimer of FKBP25 and FRB in Live Cells
Acs Cent.Sci., 2, 2016
|
|
6OYH
 
 | | Crystal structure of MraY bound to carbacaprazamycin | | Descriptor: | (5S)-5'-O-(5-amino-5-deoxy-beta-D-ribofuranosyl)-5'-C-[(2S,5S,6S)-5-carboxy-6-heptadecyl-1,4-dimethyl-3-oxo-1,4-diazepan-2-yl]uridine, MraYAA nanobody, Phospho-N-acetylmuramoyl-pentapeptide-transferase | | Authors: | Mashalidis, E.H, Lee, S.Y. | | Deposit date: | 2019-05-14 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Chemical logic of MraY inhibition by antibacterial nucleoside natural products.
Nat Commun, 10, 2019
|
|
6OYZ
 
 | | Crystal structure of MraY bound to capuramycin | | Descriptor: | (2~{S},3~{S},4~{S})-2-[(1~{R})-2-azanyl-1-[(2~{S},3~{S},4~{R},5~{R})-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3-methoxy-4-oxidanyl-oxolan-2-yl]-2-oxidanylidene-ethoxy]-3,4-bis(oxidanyl)-~{N}-[(3~{S})-2-oxidanylideneazepan-3-yl]-3,4-dihydro-2~{H}-pyran-6-carboxamide, MraYAA nanobody, Phospho-N-acetylmuramoyl-pentapeptide-transferase | | Authors: | Mashalidis, E.H, Lee, S.Y. | | Deposit date: | 2019-05-15 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.62 Å) | | Cite: | Chemical logic of MraY inhibition by antibacterial nucleoside natural products.
Nat Commun, 10, 2019
|
|
6OZ6
 
 | | Crystal structure of MraY bound to 3'-hydroxymureidomycin A | | Descriptor: | (2~{S})-2-[[(2~{S})-1-[[(2~{S},3~{S})-3-[[(2~{S})-2-azanyl-3-(3-hydroxyphenyl)propanoyl]-methyl-amino]-1-[[(~{Z})-[(3~{S},4~{R},5~{R})-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-ylidene]methyl]amino]-1-oxidanylidene-butan-2-yl]amino]-4-methylsulfanyl-1-oxidanylidene-butan-2-yl]carbamoylamino]-3-(3-hydroxyphenyl)propanoic acid, MraYAA nanobody, Phospho-N-acetylmuramoyl-pentapeptide-transferase | | Authors: | Mashalidis, E.H, Lee, S.Y. | | Deposit date: | 2019-05-15 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Chemical logic of MraY inhibition by antibacterial nucleoside natural products.
Nat Commun, 10, 2019
|
|
8TZI
 
 | | Human equilibrative nucleoside transporter-1, JH-ENT-01 bound | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3-[4-[3-[3,4-dimethoxy-5-[[4-[oxidanyl(oxidanylidene)-$l^{4}-azanyl]phenyl]methoxy]phenyl]carbonyloxypropyl]-1,4-diazepan-1-yl]propyl 3,4,5-trimethoxybenzoate, Equilibrative nucleoside transporter 1 | | Authors: | Wright, N.J, Lee, S.Y. | | Deposit date: | 2023-08-26 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Rational ENT1 inhibition as a pathway for neuropathic pain relief
To Be Published
|
|
8CXR
 
 | | Crystal structure of MraY bound to a sphaerimicin analogue | | Descriptor: | (1S,4R,5S,6R,7S,9S,10S,11S,13S,14R)-9-[(2S,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxyoxolan-2-yl]-14-(hexadecanoyloxy)-5,6,13-trihydroxy-8,16-dioxa-2,11-diazatricyclo[9.3.1.1~4,7~]hexadecane-10-carboxylic acid, MraYAA nanobody, Phospho-N-acetylmuramoyl-pentapeptide-transferase | | Authors: | Mashalidis, E.H, Lee, S.Y. | | Deposit date: | 2022-05-22 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Synthesis of macrocyclic nucleoside antibacterials and their interactions with MraY.
Nat Commun, 13, 2022
|
|
4DCK
 
 | | Crystal structure of the C-terminus of voltage-gated sodium channel in complex with FGF13 and CaM | | Descriptor: | Calmodulin, Fibroblast growth factor 13, MAGNESIUM ION, ... | | Authors: | Chung, B.C, Wang, C, Yan, H, Pitt, G.S, Lee, S.Y. | | Deposit date: | 2012-01-17 | | Release date: | 2012-06-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Ternary Complex of a NaV C-Terminal Domain, a Fibroblast Growth Factor Homologous Factor, and Calmodulin.
Structure, 20, 2012
|
|
6E0K
 
 | | Structure of Rhodothermus marinus CdnE c-UMP-AMP synthase | | Descriptor: | cGAS/DncV-like nucleotidyltransferase in E. coli homolog | | Authors: | Eaglesham, J.B, Whiteley, A.T, de Oliveira Mann, C.C, Morehouse, B.R, Nieminen, E.A, King, D.S, Lee, A.S.Y, Mekalanos, J.J, Kranzusch, P.J. | | Deposit date: | 2018-07-06 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Bacterial cGAS-like enzymes synthesize diverse nucleotide signals.
Nature, 567, 2019
|
|
