3LCS
 
 | |
4ZB1
 
 | | Crystal Structure of Blue Chromoprotein sgBP from Stichodactyla Gigantea | | Descriptor: | 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, Blue chromoprotein, sgBP | | Authors: | Lee, C.C, Ching, C.Y, Tsai, H.J, Wang, A.H.J. | | Deposit date: | 2015-04-14 | | Release date: | 2015-08-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Chromophore Deprotonation State Alters the Optical Properties of Blue Chromoprotein
Plos One, 10, 2015
|
|
2LKP
 
 | | solution structure of apo-NmtR | | Descriptor: | HTH-type transcriptional regulator NmtR | | Authors: | Lee, C, Giedroc, D. | | Deposit date: | 2011-10-18 | | Release date: | 2012-04-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Mycobacterium tuberculosis NmtR in the Apo State: Insights into Ni(II)-Mediated Allostery.
Biochemistry, 51, 2012
|
|
2Z9L
 
 | |
2Z9J
 
 | | Complex structure of SARS-CoV 3C-like protease with EPDTC | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, zinc(II)hydrogensulfide | | Authors: | Lee, C.C, Wang, A.H. | | Deposit date: | 2007-09-20 | | Release date: | 2007-12-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis of mercury- and zinc-conjugated complexes as SARS-CoV 3C-like protease inhibitors.
Febs Lett., 581, 2007
|
|
6ABT
 
 | | Crystal structure of transcription factor from Listeria monocytogenes | | Descriptor: | PadR family transcriptional regulator | | Authors: | Lee, C, Hong, M. | | Deposit date: | 2018-07-23 | | Release date: | 2019-06-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based molecular characterization and regulatory mechanism of the LftR transcription factor from Listeria monocytogenes: Conformational flexibilities and a ligand-induced regulatory mechanism.
Plos One, 14, 2019
|
|
6ABQ
 
 | | Crystal structure of transcription factor from Listeria monocytogenes | | Descriptor: | CHLORIDE ION, PadR family transcriptional regulator | | Authors: | Lee, C, Hong, M. | | Deposit date: | 2018-07-23 | | Release date: | 2019-06-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based molecular characterization and regulatory mechanism of the LftR transcription factor from Listeria monocytogenes: Conformational flexibilities and a ligand-induced regulatory mechanism.
Plos One, 14, 2019
|
|
5VRE
 
 | |
2Z9K
 
 | | Complex structure of SARS-CoV 3C-like protease with JMF1600 | | Descriptor: | (dimethylamino)(hydroxy)zinc', 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Lee, C.C, Wang, A.H. | | Deposit date: | 2007-09-20 | | Release date: | 2007-12-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis of mercury- and zinc-conjugated complexes as SARS-CoV 3C-like protease inhibitors.
Febs Lett., 581, 2007
|
|
2L14
 
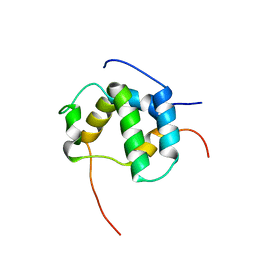 | | Structure of CBP nuclear coactivator binding domain in complex with p53 TAD | | Descriptor: | CREB-binding protein, Cellular tumor antigen p53 | | Authors: | Lee, C, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2010-07-22 | | Release date: | 2010-11-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the p53 transactivation domain in complex with the nuclear receptor coactivator binding domain of CREB binding protein.
Biochemistry, 49, 2010
|
|
2ETM
 
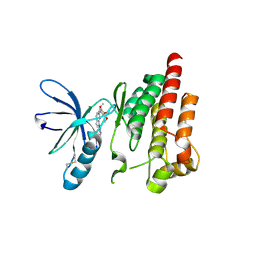 | |
3TNU
 
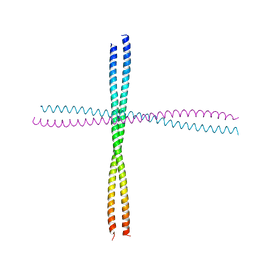 | | Heterocomplex of coil 2B domains of human intermediate filament proteins, keratin 5 (KRT5) and keratin 14 (KRT14) | | Descriptor: | Keratin, type I cytoskeletal 14, type II cytoskeletal 5 | | Authors: | Lee, C.H, Kim, M.S, Leahy, D.J, Coulombe, P.A. | | Deposit date: | 2011-09-02 | | Release date: | 2012-06-20 | | Last modified: | 2012-08-01 | | Method: | X-RAY DIFFRACTION (3.005 Å) | | Cite: | Structural basis for heteromeric assembly and perinuclear organization of keratin filaments.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3NOW
 
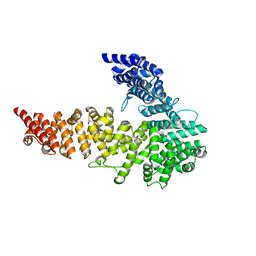 | | UNC-45 from Drosophila melanogaster | | Descriptor: | UNC-45 protein, SD10334p | | Authors: | Lee, C.F, Hauenstein, A.V, Fleming, J.K, Gasper, W.C, Engelke, V, Banumathi, S, Bernstein, S.I, Huxford, T. | | Deposit date: | 2010-06-25 | | Release date: | 2011-03-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.992 Å) | | Cite: | X-ray Crystal Structure of the UCS Domain-Containing UNC-45 Myosin Chaperone from Drosophila melanogaster.
Structure, 19, 2011
|
|
2Z9G
 
 | | Complex structure of SARS-CoV 3C-like protease with PMA | | Descriptor: | 3C-like proteinase, BENZENE, MERCURY (II) ION | | Authors: | Lee, C.C, Wang, A.H. | | Deposit date: | 2007-09-19 | | Release date: | 2007-12-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural basis of mercury- and zinc-conjugated complexes as SARS-CoV 3C-like protease inhibitors.
Febs Lett., 581, 2007
|
|
5HXN
 
 | | Crystal Structure of Z,Z-Farnesyl Diphosphate Synthase (D71M and E75A mutants) from the Wild Tomato Solanum habrochaites | | Descriptor: | (2Z,6Z)-farnesyl diphosphate synthase, chloroplastic | | Authors: | Lee, C.C, Chan, Y.T, Wang, A.H.J. | | Deposit date: | 2016-01-31 | | Release date: | 2017-04-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure and Potential Head-to-Middle Condensation Function of aZ,Z-Farnesyl Diphosphate Synthase.
Acs Omega, 2, 2017
|
|
5HXO
 
 | | Crystal Structure of Z,Z-Farnesyl Diphosphate Synthase with D71M, E75A and H103Y Mutants | | Descriptor: | (2Z,6Z)-farnesyl diphosphate synthase, chloroplastic | | Authors: | Lee, C.C, Chan, Y.T, Wang, A.H.J. | | Deposit date: | 2016-01-31 | | Release date: | 2017-04-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure and Potential Head-to-Middle Condensation Function of aZ,Z-Farnesyl Diphosphate Synthase.
Acs Omega, 2, 2017
|
|
5HXT
 
 | | Crystal Structure of Z,Z-Farnesyl Diphosphate Synthase (D71M, E75A and H103Y Mutants) Complexed with IPP and DMSPP | | Descriptor: | (2Z,6Z)-farnesyl diphosphate synthase, chloroplastic, 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, ... | | Authors: | Lee, C.C, Chan, Y.T, Wang, A.H.J. | | Deposit date: | 2016-01-31 | | Release date: | 2017-04-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure and Potential Head-to-Middle Condensation Function of a Z,Z-Farnesyl Diphosphate Synthase
Acs Omega, 2, 2017
|
|
4PX6
 
 | | SYK catalytic domain in complex with a potent pyridopyrimidinone inhibitor | | Descriptor: | 7-{[(1R,2S)-2-aminocyclohexyl]amino}-5-(1H-indol-7-ylamino)pyrido[4,3-d]pyrimidin-4(3H)-one, Tyrosine-protein kinase SYK | | Authors: | Lee, C.C. | | Deposit date: | 2014-03-21 | | Release date: | 2014-04-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Syk inhibitors with high potency in presence of blood.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
3MKQ
 
 | |
3MKR
 
 | |
5CY3
 
 | |
2HIW
 
 | |
2Z94
 
 | | Complex structure of SARS-CoV 3C-like protease with TDT | | Descriptor: | 4-methylbenzene-1,2-dithiol, Replicase polyprotein 1ab, ZINC ION | | Authors: | Lee, C.C, Wang, A.H. | | Deposit date: | 2007-09-17 | | Release date: | 2007-12-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural basis of mercury- and zinc-conjugated complexes as SARS-CoV 3C-like protease inhibitors
Febs Lett., 581, 2007
|
|
2ZU3
 
 | | Complex structure of CVB3 3C protease with TG-0204998 | | Descriptor: | 3C proteinase, N-[(benzyloxy)carbonyl]-3-[(2,2-dimethylpropanoyl)amino]-L-alanyl-N-[(1R)-4-oxo-1-{[(3S)-2-oxopyrrolidin-3-yl]methyl}pentyl]-L-leucinamide | | Authors: | Lee, C.C, Tsui, Y.C, Wang, A.H.-J. | | Deposit date: | 2008-10-12 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis of Inhibition Specificities of 3C and 3C-like Proteases by Zinc-coordinating and Peptidomimetic Compounds
J.Biol.Chem., 284, 2009
|
|
2ZTX
 
 | | Complex structure of CVB3 3C protease with EPDTC | | Descriptor: | 3C proteinase, zinc(II)hydrogensulfide | | Authors: | Lee, C.C, Wang, A.H.-J. | | Deposit date: | 2008-10-10 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural Basis of Inhibition Specificities of 3C and 3C-like Proteases by Zinc-coordinating and Peptidomimetic Compounds
J.Biol.Chem., 284, 2009
|
|
