3T03
 
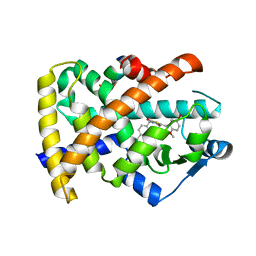 | | Crystal structure of PPAR gamma ligand binding domain in complex with a novel partial agonist GQ-16 | | Descriptor: | (5Z)-5-(5-bromo-2-methoxybenzylidene)-3-(4-methylbenzyl)-1,3-thiazolidine-2,4-dione, Nuclear receptor coactivator 1, Peroxisome proliferator-activated receptor gamma | | Authors: | Rajagopalan, S, Webb, P, Baxter, J.D, Brennan, R.G, Phillips, K.J. | | Deposit date: | 2011-07-19 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | GQ-16, a novel peroxisome proliferator-activated receptor (PPAR gamma) ligand, promotes insulin sensitization without weight gain.
J.Biol.Chem., 287, 2012
|
|
8TN8
 
 | |
2GAB
 
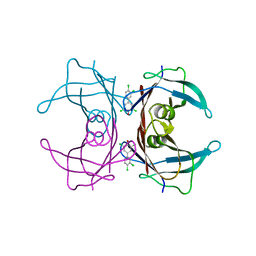 | | Human Transthyretin (TTR) Complexed with Hydroxylated polychlorinated Biphenyl-4-hydroxy-3,3',5,4'-tetrachlorobiphenyl | | Descriptor: | 3,3',4',5-TETRACHLOROBIPHENYL-4-OL, Transthyretin | | Authors: | Palaninathan, S.K, Smith, C, Safe, S.H, Kelly, J.W, Sacchettini, J.C. | | Deposit date: | 2006-03-08 | | Release date: | 2006-03-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Hydroxylated polychlorinated biphenyls selectively bind transthyretin in blood and inhibit amyloidogenesis: rationalizing rodent PCB toxicity
Chem.Biol., 11, 2004
|
|
2F8I
 
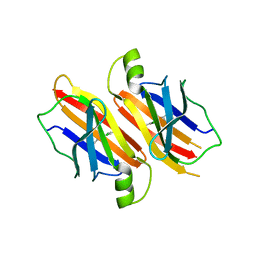 | | Human transthyretin (TTR) complexed with Benzoxazole | | Descriptor: | 2-(2,6-DICHLOROPHENYL)-1,3-BENZOXAZOLE-6-CARBOXYLIC ACID, Transthyretin | | Authors: | Palaninathan, S.K, Mohamedmohaideen, N.N, Sacchettini, J.C, Kelly, J.W. | | Deposit date: | 2005-12-02 | | Release date: | 2005-12-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.541 Å) | | Cite: | Benzoxazoles as transthyretin amyloid fibril inhibitors: synthesis, evaluation, and mechanism of action
Angew.Chem.Int.Ed.Engl., 42, 2003
|
|
7NPO
 
 | | Branched K48-K63-Ub3 | | Descriptor: | GLYCEROL, Polyubiquitin-B | | Authors: | Lange, S.M, Kwasna, D, Kulathu, Y. | | Deposit date: | 2021-02-27 | | Release date: | 2022-08-10 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | VCP/p97-associated proteins are binders and debranching enzymes of K48-K63-branched ubiquitin chains.
Nat.Struct.Mol.Biol., 2024
|
|
6KQC
 
 | |
6KP5
 
 | |
4V4M
 
 | | 1.45 Angstrom Structure of STNV coat protein | | Descriptor: | CALCIUM ION, Coat protein | | Authors: | Lane, S.W, Dennis, C.A, Lane, C.L, Trinh, C.H, Rizkallah, P.J, Stockley, P.G, Phillips, S.E.V. | | Deposit date: | 2011-04-28 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Construction and crystal structure of recombinant STNV capsids.
J.Mol.Biol., 413, 2011
|
|
5JGL
 
 | | Crystal structure of GtmA in complex with S-Adenosylmethionine | | Descriptor: | S-ADENOSYLMETHIONINE, SODIUM ION, UbiE/COQ5 family methyltransferase, ... | | Authors: | Dolan, S.K, Bock, T, Hering, V, Jones, G.W, Blankenfeldt, W, Doyle, S. | | Deposit date: | 2016-04-20 | | Release date: | 2017-03-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural, mechanistic and functional insight into gliotoxinbis-thiomethylation inAspergillus fumigatus.
Open Biol, 7, 2017
|
|
5JGK
 
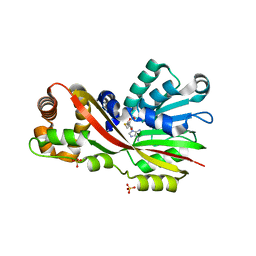 | | Crystal structure of GtmA in complex with SAH | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, UbiE/COQ5 family methyltransferase, ... | | Authors: | Dolan, S.K, Bock, T, Hering, V, Jones, G.W, Blankenfeldt, W, Dolye, S. | | Deposit date: | 2016-04-20 | | Release date: | 2017-03-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Structural, mechanistic and functional insight into gliotoxinbis-thiomethylation inAspergillus fumigatus.
Open Biol, 7, 2017
|
|
8S9L
 
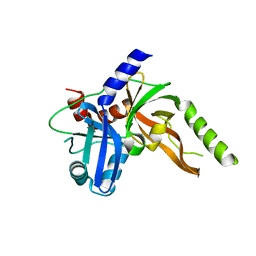 | | Structure of monomeric FAM111A SPD V347D Mutant | | Descriptor: | SULFATE ION, Serine protease FAM111A | | Authors: | Palani, S, Alvey, J.A, Cong, A.T.Q, Schellenberg, M.J, Machida, Y. | | Deposit date: | 2023-03-29 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Dimerization-dependent serine protease activity of FAM111A prevents replication fork stalling at topoisomerase 1 cleavage complexes.
Nat Commun, 15, 2024
|
|
5JGJ
 
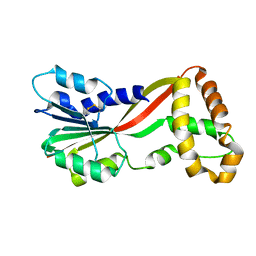 | | Crystal structure of GtmA | | Descriptor: | UbiE/COQ5 family methyltransferase, putative | | Authors: | Dolan, S.K, Bock, T, Hering, V, Jones, G.W, Blankenfeldt, W, Doyle, S. | | Deposit date: | 2016-04-20 | | Release date: | 2017-03-01 | | Last modified: | 2018-03-28 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural, mechanistic and functional insight into gliotoxinbis-thiomethylation inAspergillus fumigatus.
Open Biol, 7, 2017
|
|
7TMW
 
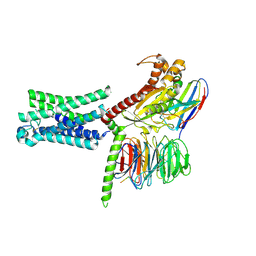 | | Cryo-EM structure of the relaxin receptor RXFP1 in complex with heterotrimeric Gs | | Descriptor: | Camelid antibody VHH fragment Nb35, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Erlandson, S.C, Rawson, S, Kruse, A.C. | | Deposit date: | 2022-01-20 | | Release date: | 2023-02-15 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The relaxin receptor RXFP1 signals through a mechanism of autoinhibition.
Nat.Chem.Biol., 19, 2023
|
|
7RGG
 
 | | Room temperature serial crystal structure of Glutaminase C in complex with inhibitor BPTES | | Descriptor: | Glutaminase kidney isoform, mitochondrial 68 kDa chain, N,N'-[sulfanediylbis(ethane-2,1-diyl-1,3,4-thiadiazole-5,2-diyl)]bis(2-phenylacetamide) | | Authors: | Milano, S.K, Finke, A, Cerione, R.A. | | Deposit date: | 2021-07-15 | | Release date: | 2022-05-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | New insights into the molecular mechanisms of glutaminase C inhibitors in cancer cells using serial room temperature crystallography.
J.Biol.Chem., 298, 2022
|
|
7REN
 
 | | Room temperature serial crystal structure of Glutaminase C in complex with inhibitor UPGL-00004 | | Descriptor: | 2-phenyl-N-{5-[4-({5-[(phenylacetyl)amino]-1,3,4-thiadiazol-2-yl}amino)piperidin-1-yl]-1,3,4-thiadiazol-2-yl}acetamide, Glutaminase kidney isoform, mitochondrial | | Authors: | Milano, S.K, Finke, A, Cerione, R.A. | | Deposit date: | 2021-07-13 | | Release date: | 2022-05-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | New insights into the molecular mechanisms of glutaminase C inhibitors in cancer cells using serial room temperature crystallography.
J.Biol.Chem., 298, 2022
|
|
8S9K
 
 | | Structure of dimeric FAM111A SPD S541A Mutant | | Descriptor: | GLYCEROL, Serine protease FAM111A | | Authors: | Palani, S, Alvey, J.A, Cong, A.T.Q, Schellenberg, M.J, Machida, Y. | | Deposit date: | 2023-03-29 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Dimerization-dependent serine protease activity of FAM111A prevents replication fork stalling at topoisomerase 1 cleavage complexes.
Nat Commun, 15, 2024
|
|
7NPI
 
 | | Crystal structure of Mindy2 (C266A) in complex with Lys48-linked penta-ubiquitin (K48-Ub5) | | Descriptor: | CHLORIDE ION, Polyubiquitin-C, SODIUM ION, ... | | Authors: | Lange, S.M, Armstrong, L.A, Kulathu, Y. | | Deposit date: | 2021-02-26 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Mechanism of activation and regulation of deubiquitinase activity in MINDY1 and MINDY2.
Mol.Cell, 81, 2021
|
|
4QJY
 
 | | Crystal structure of native Ara127N, a GH127 beta-L-arabinofuranosidase from Geobacillus Stearothermophilus T6 | | Descriptor: | ACETATE ION, GH127 beta-L-arabinofuranosidase | | Authors: | Lansky, S, Salama, R, Dann, R, Shner, I, Manjasetty, B, Belrhali, H, Shoham, Y, Shoham, G. | | Deposit date: | 2014-06-05 | | Release date: | 2015-06-10 | | Method: | X-RAY DIFFRACTION (2.294 Å) | | Cite: | Crystal structure of native Ara127N, a GH127 beta-L-arabinofuranosidase from Geobacillus Stearothermophilus T6
To be Published
|
|
5XTU
 
 | | Crystal Structure of GDSL Esterase of Photobacterium sp. J15 | | Descriptor: | 1,2-ETHANEDIOL, CACODYLATE ION, CALCIUM ION, ... | | Authors: | Mazlan, S.N.H.S, Jonet, M.A, Leow, T.C, Ali, M.S.M, Rahman, R.N.Z.R.A. | | Deposit date: | 2017-06-21 | | Release date: | 2018-10-10 | | Last modified: | 2018-10-17 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Crystallization and structure elucidation of GDSL esterase of Photobacterium sp. J15.
Int. J. Biol. Macromol., 119, 2018
|
|
8A67
 
 | |
5WKZ
 
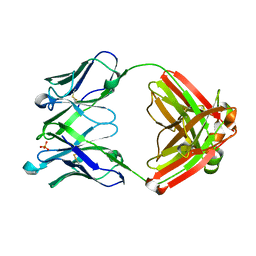 | | VH1-69 germline antibody predicted from CR6261 | | Descriptor: | Immunoglobulin heavy variable 1-69D,IgG H chain, Lambda-chain (AA -20 to 215), SULFATE ION | | Authors: | Lang, S, Lee, P.S. | | Deposit date: | 2017-07-25 | | Release date: | 2018-08-01 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Anti-idiotypic antibody K1-18 engages VH1-69 precursor and affinity-matured, anti-stem antibodies through mimicry of the HA stem
To Be Published
|
|
1L2W
 
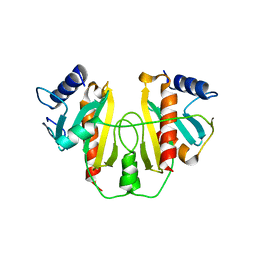 | |
1YDV
 
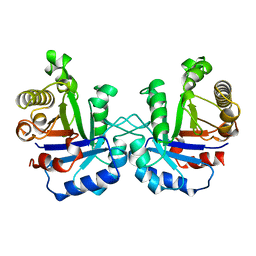 | | TRIOSEPHOSPHATE ISOMERASE (TIM) | | Descriptor: | TRIOSEPHOSPHATE ISOMERASE | | Authors: | Velankar, S.S, Murthy, M.R.N. | | Deposit date: | 1997-04-24 | | Release date: | 1997-10-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Triosephosphate isomerase from Plasmodium falciparum: the crystal structure provides insights into antimalarial drug design.
Structure, 5, 1997
|
|
4QK0
 
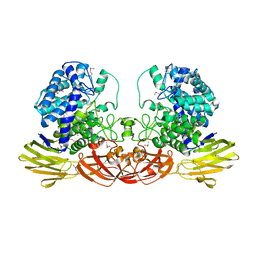 | | Crystal structure of Ara127N-Se, a GH127 beta-L-arabinofuranosidase from Geobacillus Stearothermophilus T6 | | Descriptor: | GH127 beta-L-arabinofuranoside | | Authors: | Lansky, S, Salama, R, Dann, R, Shner, I, Manjasetty, B, Belrhali, H, Shoham, Y, Shoham, G. | | Deposit date: | 2014-06-05 | | Release date: | 2015-06-10 | | Method: | X-RAY DIFFRACTION (2.258 Å) | | Cite: | Crystal structure of Ara127N-Se, a GH127 beta-L-arabinofuranosidase from Geobacillus Stearothermophilus T6
To be Published
|
|
8GKA
 
 | | Human TRPV3 tetramer structure, closed conformation | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, SODIUM ION, Transient receptor potential cation channel subfamily V member 3 | | Authors: | Lansky, S, Betancourt, J.M, Scheuring, S. | | Deposit date: | 2023-03-17 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | A pentameric TRPV3 channel with a dilated pore.
Nature, 621, 2023
|
|
