6AR0
 
 | | Structure of human SLMAP FHA domain | | Descriptor: | Sarcolemmal membrane-associated protein | | Authors: | Ni, L, Luo, X. | | Deposit date: | 2017-08-21 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | SAV1 promotes Hippo kinase activation through antagonizing the PP2A phosphatase STRIPAK.
Elife, 6, 2017
|
|
6AR2
 
 | |
3GMH
 
 | | Crystal Structure of the Mad2 Dimer | | Descriptor: | Mitotic spindle assembly checkpoint protein MAD2A, SULFATE ION | | Authors: | Ozkan, E, Luo, X, Machius, M, Yu, H, Deisenhofer, J. | | Deposit date: | 2009-03-13 | | Release date: | 2010-11-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.95 Å) | | Cite: | Structure of an intermediate conformer of the spindle checkpoint protein Mad2.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3K3E
 
 | | Crystal structure of the PDE9A catalytic domain in complex with (R)-BAY73-6691 | | Descriptor: | 1-(2-chlorophenyl)-6-[(2R)-3,3,3-trifluoro-2-methylpropyl]-1,7-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, ... | | Authors: | Wang, H, Luo, X, Ye, M, Hou, J, Robinson, H, Ke, H. | | Deposit date: | 2009-10-02 | | Release date: | 2010-02-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Insight into Binding of Phosphodiesterase-9A Selective Inhibitors by Crystal Structures and Mutagenesis
J.Med.Chem., 53, 2010
|
|
3K3H
 
 | | Crystal structure of the PDE9A catalytic domain in complex with (S)-BAY73-6691 | | Descriptor: | 1-(2-chlorophenyl)-6-[(2S)-3,3,3-trifluoro-2-methylpropyl]-1,7-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, ... | | Authors: | Wang, H, Luo, X, Ye, M, Hou, J, Robinson, H, Ke, H. | | Deposit date: | 2009-10-02 | | Release date: | 2010-02-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insight into Binding of Phosphodiesterase-9A Selective Inhibitors by Crystal Structures and Mutagenesis
J.Med.Chem., 53, 2010
|
|
4QQN
 
 | | Protein arginine methyltransferase 3 in complex with compound MTV044246 | | Descriptor: | 1-{2-[1-(aminomethyl)cyclohexyl]ethyl}-3-isoquinolin-6-ylurea, CHLORIDE ION, GLYCEROL, ... | | Authors: | Dong, A, Dobrovetsky, E, Tempel, W, He, H, Zhao, K, Smil, D, Landon, M, Luo, X, Chen, Z, Dai, M, Yu, Z, Lin, Y, Zhang, H, Zhao, K, Schapira, M, Brown, P.J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Vedadi, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-27 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Discovery of Potent and Selective Allosteric Inhibitors of Protein Arginine Methyltransferase 3 (PRMT3).
J. Med. Chem., 61, 2018
|
|
6LO2
 
 | | Crystal structure of EED in complex with EZH2 peptide and compound 11# | | Descriptor: | 8-[4-[(dimethylamino)methyl]phenyl]-N-[(2-methoxyphenyl)methyl]-[1,2,4]triazolo[4,3-a]pyridin-5-amine, Histone-lysine N-methyltransferase EZH2, Polycomb protein EED | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H, Chen, Z. | | Deposit date: | 2020-01-02 | | Release date: | 2021-07-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure of EED in complex with EZH2 peptide and cmpd20
To Be Published
|
|
6OCG
 
 | | Crystal structure of VASH1-SVBP complex bound with EpoY | | Descriptor: | CHLORIDE ION, GLYCEROL, N-[(3R)-4-ethoxy-3-hydroxy-4-oxobutanoyl]-L-tyrosine, ... | | Authors: | Li, F, Luo, X, Yu, H. | | Deposit date: | 2019-03-23 | | Release date: | 2019-06-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.833 Å) | | Cite: | Structural basis of tubulin detyrosination by vasohibins.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6OCF
 
 | | The crystal structure of VASH1-SVBP complex | | Descriptor: | CHLORIDE ION, GLYCEROL, Small vasohibin-binding protein, ... | | Authors: | Li, F, Luo, X, Yu, H. | | Deposit date: | 2019-03-23 | | Release date: | 2019-06-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Structural basis of tubulin detyrosination by vasohibins.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6OCH
 
 | | Crystal structure of VASH1-SVBP complex bound with parthenolide | | Descriptor: | GLYCEROL, SULFATE ION, Small vasohibin-binding protein, ... | | Authors: | Li, F, Luo, X, Yu, H. | | Deposit date: | 2019-03-23 | | Release date: | 2019-06-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Structural basis of tubulin detyrosination by vasohibins.
Nat.Struct.Mol.Biol., 26, 2019
|
|
2IW5
 
 | | Structural Basis for CoREST-Dependent Demethylation of Nucleosomes by the Human LSD1 Histone Demethylase | | Descriptor: | AMMONIUM ION, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Yang, M, Gocke, C.B, Luo, X, Borek, D, Tomchick, D.R, Machius, M, Otwinowski, Z, Yu, H. | | Deposit date: | 2006-06-26 | | Release date: | 2006-08-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural Basis for Corest-Dependent Demethylation of Nucleosomes by the Human Lsd1 Histone Demethylase
Mol.Cell, 23, 2006
|
|
8X8U
 
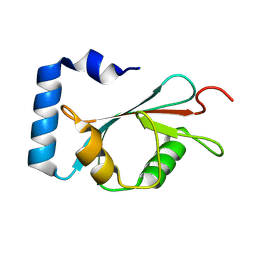 | |
2QYF
 
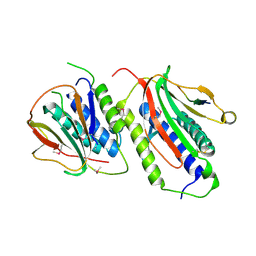 | |
5WC2
 
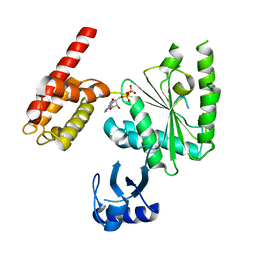 | | Crystal Structure of ADP-bound human TRIP13 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Pachytene checkpoint protein 2 homolog | | Authors: | Jeong, B.-C, Luo, X. | | Deposit date: | 2017-06-29 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanistic insight into TRIP13-catalyzed Mad2 structural transition and spindle checkpoint silencing.
Nat Commun, 8, 2017
|
|
2VFX
 
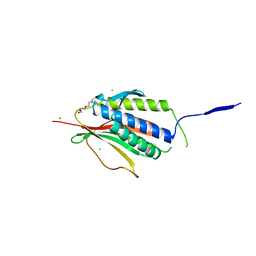 | | Structure of the Symmetric Mad2 Dimer | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, CHLORIDE ION, ... | | Authors: | Yang, M, Li, B, Liu, C.-J, Tomchick, D.R, Machius, M, Rizo, J, Yu, H, Luo, X. | | Deposit date: | 2007-11-05 | | Release date: | 2008-03-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Insights Into MAD2 Regulation in the Spindle Checkpoint Revealed by the Crystal Structure of the Symmetric MAD2 Dimer.
Plos Biol., 6, 2008
|
|
