4O6O
 
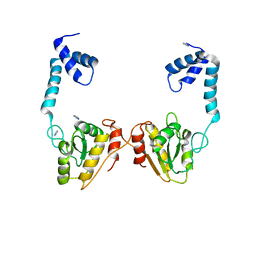 | | Structural and functional studies the characterization of Cys4 Zinc-finger motif in the recombination mediator protein RecR | | Descriptor: | IMIDAZOLE, Recombination protein RecR, ZINC ION | | Authors: | Tang, Q, Liu, Y.P, Yan, X.X, Liang, D.C. | | Deposit date: | 2013-12-23 | | Release date: | 2014-12-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and functional characterization of Cys4 zinc finger motif in the recombination mediator protein RecR.
DNA Repair (Amst.), 24, 2014
|
|
4E9U
 
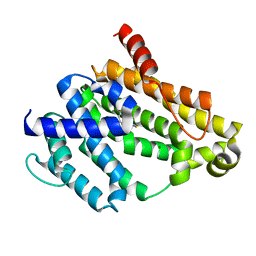 | | Crystal structure of dehydrosqualene synthase (Crtm) from S. aureus complexed with a thiocyanate inhibitor | | Descriptor: | 2-(4-phenoxyphenoxy)ethyl thiocyanate, Dehydrosqualene synthase, MAGNESIUM ION | | Authors: | Lin, F.-Y, Axelson, J, Liu, Y.-L, Zhnag, Y, Oldfield, E. | | Deposit date: | 2012-03-21 | | Release date: | 2012-04-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Head-to-Head Prenyl Tranferases: Anti-Infective Drug Targets.
J.Med.Chem., 55, 2012
|
|
5FWM
 
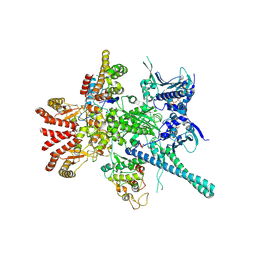 | | Atomic cryoEM structure of Hsp90-Cdc37-Cdk4 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CYCLIN-DEPENDENT KINASE 4, HEAT SHOCK PROTEIN HSP 90 BETA, ... | | Authors: | Verba, K.A, Wang, R.Y.R, Arakawa, A, Liu, Y, Yokoyama, S, Agard, D.A. | | Deposit date: | 2016-02-18 | | Release date: | 2016-07-06 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Atomic Structure of Hsp90-Cdc37-Cdk4 Reveals that Hsp90 Traps and Stabilizes an Unfolded Kinase.
Science, 352, 2016
|
|
2N3J
 
 | | Solution Structure of the alpha-crystallin domain from the redox-sensitive chaperone, HSPB1 | | Descriptor: | Heat shock protein beta-1 | | Authors: | Rajagopal, P, Liu, Y, Shi, L, Klevit, R.E. | | Deposit date: | 2015-06-03 | | Release date: | 2015-08-19 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structure of the alpha-crystallin domain from the redox-sensitive chaperone, HSPB1.
J.Biomol.Nmr, 63, 2015
|
|
4O6P
 
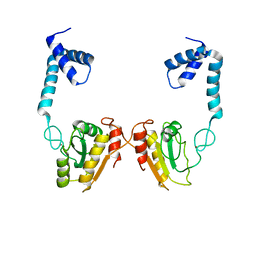 | | Structural and functional studies the characterization of C58G/C70G mutant in Cys4 Zinc-finger motif in the recombination mediator protein RecR | | Descriptor: | Recombination protein RecR, ZINC ION | | Authors: | Tang, Q, Liu, Y.P, Yan, X.X, Liang, D.C. | | Deposit date: | 2013-12-23 | | Release date: | 2014-12-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and functional characterization of Cys4 zinc finger motif in the recombination mediator protein RecR.
DNA Repair (Amst.), 24, 2014
|
|
2OIG
 
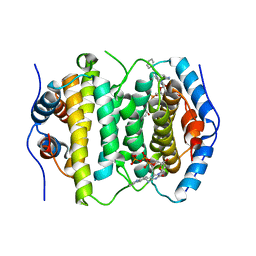 | | Crystal structure of RS21-C6 core segment and dm5CTP complex | | Descriptor: | 2'-DEOXY-5-METHYLCYTIDINE 5'-(TETRAHYDROGEN TRIPHOSPHATE), RS21-C6 | | Authors: | Wu, B, Liu, Y, Zhao, Q, Liao, S, Zhang, J, Bartlam, M, Chen, W, Rao, Z. | | Deposit date: | 2007-01-11 | | Release date: | 2007-03-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal Structure of RS21-C6, Involved in Nucleoside Triphosphate Pyrophosphohydrolysis
J.Mol.Biol., 367, 2007
|
|
1C78
 
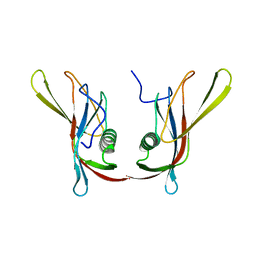 | | STAPHYLOKINASE (SAK) DIMER | | Descriptor: | STAPHYLOKINASE | | Authors: | Rao, Z, Jiang, F, Liu, Y, Zhang, X, Chen, Y, Bartlam, M, Song, H, Ding, Y. | | Deposit date: | 2000-02-01 | | Release date: | 2000-08-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a staphylokinase: variant a model for reduced antigenicity.
Eur.J.Biochem., 269, 2002
|
|
1C77
 
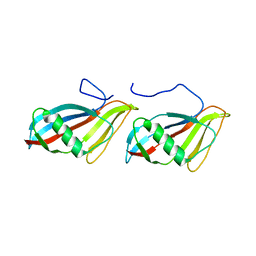 | | STAPHYLOKINASE (SAK) DIMER | | Descriptor: | STAPHYLOKINASE | | Authors: | Rao, Z, Jiang, F, Liu, Y, Zhang, X, Chen, Y, Bartlam, M, Song, H, Ding, Y. | | Deposit date: | 2000-02-01 | | Release date: | 2000-08-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a staphylokinase: variant a model for reduced antigenicity.
Eur.J.Biochem., 269, 2002
|
|
1C79
 
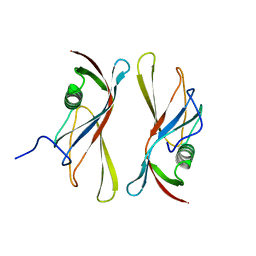 | | STAPHYLOKINASE (SAK) DIMER | | Descriptor: | STAPHYLOKINASE | | Authors: | Rao, Z, Jiang, F, Liu, Y, Zhang, X, Chen, Y, Bartlam, M, Song, H, Ding, Y. | | Deposit date: | 2000-02-01 | | Release date: | 2000-08-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a staphylokinase: variant a model for reduced antigenicity.
Eur.J.Biochem., 269, 2002
|
|
2VQ4
 
 | |
2K5U
 
 | |
2OIE
 
 | | Crystal structure of RS21-C6 core segment RSCUT | | Descriptor: | RS21-C6, SULFATE ION | | Authors: | Wu, B, Liu, Y, Zhao, Q, Liao, S, Zhang, J, Bartlam, M, Chen, W, Rao, Z. | | Deposit date: | 2007-01-10 | | Release date: | 2007-03-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of RS21-C6, Involved in Nucleoside Triphosphate Pyrophosphohydrolysis
J.Mol.Biol., 367, 2007
|
|
2O0F
 
 | | Docking of the modified RF3 X-ray structure into cryo-EM map of E.coli 70S ribosome bound with RF3 | | Descriptor: | Peptide chain release factor 3 | | Authors: | Gao, H, Zhou, Z, Rawat, U, Huang, C, Bouakaz, L, Wang, C, Liu, Y, Zavialov, A, Gursky, R, Sanyal, S, Ehrenberg, M, Frank, J, Song, H. | | Deposit date: | 2006-11-27 | | Release date: | 2007-07-24 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (15.5 Å) | | Cite: | RF3 induces ribosomal conformational changes responsible for dissociation of class I release factors
Cell(Cambridge,Mass.), 129, 2007
|
|
2LQ9
 
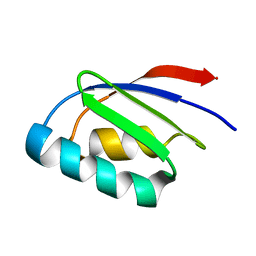 | |
3FKY
 
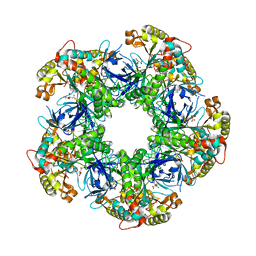 | | Crystal structure of the glutamine synthetase Gln1deltaN18 from the yeast Saccharomyces cerevisiae | | Descriptor: | CITRATE ANION, Glutamine synthetase | | Authors: | He, Y.X, Gui, L, Liu, Y.Z, Du, Y, Zhou, Y.Y, Li, P, Zhou, C.Z. | | Deposit date: | 2008-12-18 | | Release date: | 2009-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structure of Saccharomyces cerevisiae glutamine synthetase Gln1 suggests a nanotube-like supramolecular assembly
Proteins, 76, 2009
|
|
6M4Q
 
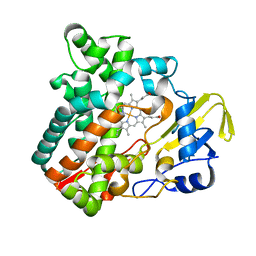 | | Cytochrome P450 monooxygenase StvP2 substrate-free structure | | Descriptor: | Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sun, G, Hu, C, Mei, Q, Luo, M, Chen, X, Li, Z, Liu, Y, Deng, Z, Zhang, Z, Sun, Y. | | Deposit date: | 2020-03-08 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Uncovering the cytochrome P450-catalyzed methylenedioxy bridge formation in streptovaricins biosynthesis.
Nat Commun, 11, 2020
|
|
5C6H
 
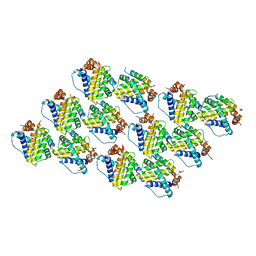 | | Mcl-1 complexed with Mule | | Descriptor: | Induced myeloid leukemia cell differentiation protein Mcl-1, Mule BH3 peptide from E3 ubiquitin-protein ligase HUWE1 | | Authors: | Song, T, Wang, Z, Ji, F, Chai, G, Liu, Y, Li, X, Li, Z, Fan, Y, Zhang, Z. | | Deposit date: | 2015-06-23 | | Release date: | 2016-08-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of Mcl-1 complexed with Mule at 2.05 Angstroms resolution
To Be Published
|
|
5H05
 
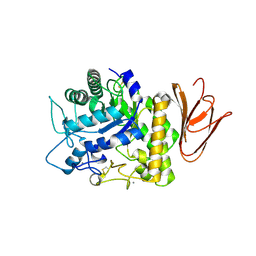 | | Crystal structure of AmyP E221Q in complex with MALTOTRIOSE | | Descriptor: | AmyP, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | He, C, Liu, Y. | | Deposit date: | 2016-10-03 | | Release date: | 2017-08-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of a raw-starch-degrading bacterial alpha-amylase belonging to subfamily 37 of the glycoside hydrolase family GH13
Sci Rep, 7, 2017
|
|
6IFE
 
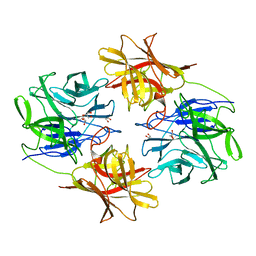 | | A Glycoside Hydrolase Family 43 beta-Xylosidase | | Descriptor: | Beta-xylosidase, GLYCEROL | | Authors: | Li, N, Liu, Y, Zhang, R, Zhou, J.P, Huang, Z.X. | | Deposit date: | 2018-09-20 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.804 Å) | | Cite: | Biochemical and structural properties of a low-temperature-active glycoside hydrolase family 43 beta-xylosidase: Activity and instability at high neutral salt concentrations.
Food Chem, 301, 2019
|
|
4F6V
 
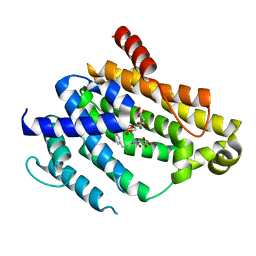 | | Crystal structure of dehydrosqualene synthase (crtm) from s. aureus complexed with bph-1034, mg2+ and fmp. | | Descriptor: | (2E,6Z)-3,7,11-trimethyldodeca-2,6,10-trien-1-yl dihydrogen phosphate, 2,4-dioxo-4-{[3-(3-phenoxyphenyl)propyl]amino}butanoic acid, Dehydrosqualene synthase, ... | | Authors: | Lin, F.-Y, Zhang, Y, Liu, Y.-L, Oldfield, E. | | Deposit date: | 2012-05-15 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | HIV-1 Integrase Inhibitor-Inspired Antibacterials Targeting Isoprenoid Biosynthesis.
ACS Med Chem Lett, 3, 2012
|
|
4F6X
 
 | | Crystal structure of dehydrosqualene synthase (crtm) from s. aureus complexed with bph-1112 | | Descriptor: | 1-[3-(hexyloxy)benzyl]-4-hydroxy-2-oxo-1,2-dihydropyridine-3-carboxylic acid, D(-)-TARTARIC ACID, Dehydrosqualene synthase, ... | | Authors: | Lin, F.-Y, Zhang, Y, Liu, Y.-L, Oldfield, E. | | Deposit date: | 2012-05-15 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | HIV-1 Integrase Inhibitor-Inspired Antibacterials Targeting Isoprenoid Biosynthesis.
ACS Med Chem Lett, 3, 2012
|
|
5GT5
 
 | |
3SGV
 
 | |
4ITI
 
 | | Crystal structure of RIP1 kinase in complex with necrostatin-3 analog | | Descriptor: | 1-{(3S,3aS)-3-[3-fluoro-4-(trifluoromethoxy)phenyl]-8-methoxy-3,3a,4,5-tetrahydro-2H-benzo[g]indazol-2-yl}-2-hydroxyethanone, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Xie, T, Peng, W, Liu, Y, Yan, C, Shi, Y. | | Deposit date: | 2013-01-18 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structural Basis of RIP1 Inhibition by Necrostatins.
Structure, 21, 2013
|
|
4ITH
 
 | | Crystal structure of RIP1 kinase in complex with necrostatin-1 analog | | Descriptor: | (5R)-5-[(7-chloro-1H-indol-3-yl)methyl]-3-methylimidazolidine-2,4-dione, IODIDE ION, Receptor-interacting serine/threonine-protein kinase 1, ... | | Authors: | Xie, T, Peng, W, Liu, Y, Yan, C, Shi, Y. | | Deposit date: | 2013-01-18 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Basis of RIP1 Inhibition by Necrostatins.
Structure, 21, 2013
|
|
