1JUD
 
 | | L-2-HALOACID DEHALOGENASE | | Descriptor: | L-2-HALOACID DEHALOGENASE | | Authors: | Hisano, T, Hata, Y, Fujii, T, Liu, J.-Q, Kurihara, T, Esaki, N, Soda, K. | | Deposit date: | 1996-06-03 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of L-2-haloacid dehalogenase from Pseudomonas sp. YL. An alpha/beta hydrolase structure that is different from the alpha/beta hydrolase fold.
J.Biol.Chem., 271, 1996
|
|
1MQA
 
 | | Crystal structure of high affinity alphaL I domain in the absence of ligand or metal | | Descriptor: | Integrin alpha-L | | Authors: | Shimaoka, T, Xiao, T, Liu, J.-H, Yang, Y, Dong, Y, Jun, C.-D, Zhang, R, Takagi, J, Wang, J.-H, Springer, T.A. | | Deposit date: | 2002-09-15 | | Release date: | 2003-01-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of the aL I domain and its complex with ICAM-1 reveal a shape-shifting pathway for integrin regulation
Cell(Cambridge,Mass.), 112, 2003
|
|
1MQ9
 
 | | Crystal structure of high affinity alphaL I domain with ligand mimetic crystal contact | | Descriptor: | Integrin alpha-L, MANGANESE (II) ION | | Authors: | Shimaoka, M, Xiao, T, Liu, J.-H, Yang, Y, Dong, Y, Jun, C.-D, McCormack, A, Zhang, R, Joachimiak, A, Takagi, J, Wang, J.-H, Springer, T.A. | | Deposit date: | 2002-09-15 | | Release date: | 2003-01-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the aL I domain and its complex with ICAM-1 reveal a shape-shifting pathway for integrin regulation
Cell(Cambridge,Mass.), 112, 2003
|
|
4IT7
 
 | | Crystal structure of Al-CPI | | Descriptor: | CPI | | Authors: | Mei, G.Q, Liu, S.L, Sun, M.Z, Liu, J. | | Deposit date: | 2013-01-17 | | Release date: | 2014-01-29 | | Last modified: | 2014-06-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for the Immunomodulatory Function of Cysteine Protease Inhibitor from Human Roundworm Ascaris lumbricoides.
Plos One, 9, 2014
|
|
3FUR
 
 | | Crystal Structure of PPARg in complex with INT131 | | Descriptor: | 2,4-dichloro-N-[3,5-dichloro-4-(quinolin-3-yloxy)phenyl]benzenesulfonamide, CHLORIDE ION, Nuclear receptor coactivator 1, ... | | Authors: | Wang, Z, Liu, J, Walker, N. | | Deposit date: | 2009-01-14 | | Release date: | 2009-06-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | INT131: a selective modulator of PPAR gamma.
J.Mol.Biol., 386, 2009
|
|
1EI8
 
 | |
8EB7
 
 | |
8EAO
 
 | |
8E4G
 
 | |
5GRJ
 
 | | Crystal structure of human PD-L1 with monoclonal antibody avelumab | | Descriptor: | Programmed cell death 1 ligand 1, avelumab H chain, avelumab L chain | | Authors: | Liu, K, Tan, S, Chai, Y, Chen, D, Song, H, Zhang, C.W.-H, Shi, Y, Liu, J, Tan, W, Lyu, J, Gao, S, Yan, J, Qi, J, Gao, G.F. | | Deposit date: | 2016-08-11 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.206 Å) | | Cite: | Structural basis of anti-PD-L1 monoclonal antibody avelumab for tumor therapy.
Cell Res., 27, 2017
|
|
4G42
 
 | | Structure of the Chicken MHC Class I Molecule BF2*0401 complexed to pepitde P8D | | Descriptor: | 8-MERIC PEPTIDE P8D, Beta-2 microglobulin, MHC class I alpha chain 2 | | Authors: | Zhang, J, Chen, Y, Qi, J, Gao, F, Liu, J, Kaufman, J, Xia, C, Gao, G.F. | | Deposit date: | 2012-07-16 | | Release date: | 2012-11-21 | | Method: | X-RAY DIFFRACTION (2.294 Å) | | Cite: | Narrow Groove and Restricted Anchors of MHC Class I Molecule BF2*0401 Plus Peptide Transporter Restriction Can Explain Disease Susceptibility of B4 Chickens.
J.Immunol., 189, 2012
|
|
7NTH
 
 | | Structure of TAK1 in complex with compound 54 | | Descriptor: | 2-[[5-[[2-[bis(fluoranyl)methoxy]phenyl]methyl-[(2~{R})-1-(methylamino)-1-oxidanylidene-propan-2-yl]carbamoyl]-1~{H}-imidazol-2-yl]carbonyl]isoindole-5-carboxamide, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Veerman, J.J.N, Bruseker, Y.B, Damen, E, Heijne, E.H, van Bruggen, W, Hekking, K.F.W, Winkel, R, Hupp, C.D, Keefe, A.D, Liu, J, Thomson, H.A, Zhang, Y, Cuozzo, J.W, McRiner, A.J, Mulvihill, M.J, van Rijnsbergen, P, Zech, B, Renzetti, L.M, Babiss, L, Mueller, G. | | Deposit date: | 2021-03-09 | | Release date: | 2021-04-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Discovery of 2,4-1 H -Imidazole Carboxamides as Potent and Selective TAK1 Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
7NTI
 
 | | Structure of TAK1 in complex with compound 22 | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, Mitogen-activated protein kinase 7,TGF-beta-activated kinase 1 and MAP3K7-binding protein 1, ... | | Authors: | Veerman, J.J.N, Bruseker, Y.B, Damen, E, Heijne, E.H, van Bruggen, W, Hekking, K.F.W, Winkel, R, Hupp, C.D, Keefe, A.D, Liu, J, Thomson, H.A, Zhang, Y, Cuozzo, J.W, McRiner, A.J, Mulvihill, M.J, van Rijnsbergen, P, Zech, B, Renzetti, L.M, Babiss, L, Mueller, G. | | Deposit date: | 2021-03-09 | | Release date: | 2021-04-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Discovery of 2,4-1 H -Imidazole Carboxamides as Potent and Selective TAK1 Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
4G43
 
 | | Structure of the chicken MHC class I molecule BF2*0401 complexed to P5E | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 8-MERIC PEPTIDE P5E, Beta-2 microglobulin, ... | | Authors: | Zhang, J, Chen, Y, Qi, J, Gao, F, Liu, J, Kaufman, J, Xia, C, Gao, G.F. | | Deposit date: | 2012-07-16 | | Release date: | 2012-11-21 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Narrow Groove and Restricted Anchors of MHC Class I Molecule BF2*0401 Plus Peptide Transporter Restriction Can Explain Disease Susceptibility of B4 Chickens.
J.Immunol., 189, 2012
|
|
1LB2
 
 | | Structure of the E. coli alpha C-terminal domain of RNA polymerase in complex with CAP and DNA | | Descriptor: | 5'-D(*CP*TP*AP*GP*AP*TP*CP*AP*CP*AP*TP*TP*TP*TP*AP*GP*GP*AP*AP*AP*AP*AP*AP*G)-3', 5'-D(*CP*TP*TP*TP*TP*TP*TP*CP*CP*TP*AP*AP*AP*AP*TP*GP*TP*GP*AP*T)-3', ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, ... | | Authors: | Benoff, B, Yang, H, Lawson, C.L, Parkinson, G, Liu, J, Blatter, E, Ebright, Y.W, Berman, H.M, Ebright, R.H. | | Deposit date: | 2002-04-01 | | Release date: | 2002-09-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis of transcription activation: the CAP-alpha CTD-DNA complex.
Science, 297, 2002
|
|
5BTU
 
 | |
1IG3
 
 | | Mouse Thiamin Pyrophosphokinase Complexed with Thiamin | | Descriptor: | 3-(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-5-(2-HYDROXY-ETHYL)-4-METHYL-THIAZOL-3-IUM, SULFATE ION, thiamin pyrophosphokinase | | Authors: | Timm, D.E, Liu, J, Baker, L.-J, Harris, R.A. | | Deposit date: | 2001-04-16 | | Release date: | 2001-04-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of thiamin pyrophosphokinase.
J.Mol.Biol., 310, 2001
|
|
6VRV
 
 | | Discovery of SARxxxx92, a pan-PIM kinase inhibitor, efficacious in a KG1 tumor model | | Descriptor: | 4-chloro-1-{(1S)-1-[(3S)-3-fluoropyrrolidin-3-yl]ethyl}-3-methyl-1H-pyrrolo[2,3-b]pyridine-6-carboxamide, GLYCEROL, Serine/threonine-protein kinase pim-1 | | Authors: | Barberis, C.E, Batchelor, J.D, Liu, J. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Discovery of SARxxxx92, a pan-PIM kinase inhibitor, efficacious in a KG1 tumor model.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
6VRU
 
 | | PIM-inhibitor complex 1 | | Descriptor: | 3,4-dichloro-2-cyclopropyl-1-[(piperidin-4-yl)methyl]-1H-pyrrolo[2,3-b]pyridine-6-carboxamide, ACETATE ION, IMIDAZOLE, ... | | Authors: | Barberis, C.E, Batchelor, J.D, Mechin, I, Liu, J. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Discovery of SARxxxx92, a pan-PIM kinase inhibitor, efficacious in a KG1 tumor model.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
7YOJ
 
 | | Structure of CasPi with guide RNA and target DNA | | Descriptor: | CasPi, DNA (30-MER), DNA (5'-D(P*CP*GP*GP*GP*AP*TP*GP*CP*CP*CP*AP*G)-3'), ... | | Authors: | Li, C.P, Wang, J, Liu, J.J. | | Deposit date: | 2022-08-01 | | Release date: | 2023-02-15 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | The compact Cas pi (Cas12l) 'bracelet' provides a unique structural platform for DNA manipulation.
Cell Res., 33, 2023
|
|
3QIC
 
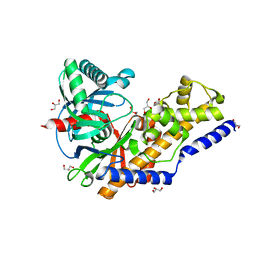 | | The structure of human glucokinase E339K mutation | | Descriptor: | GLYCEROL, Glucokinase, alpha-D-glucopyranose | | Authors: | Liu, Q, Liu, S, Liu, J. | | Deposit date: | 2011-01-27 | | Release date: | 2011-06-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of E339K mutated human glucokinase reveals changes in the ATP binding site.
Febs Lett., 585, 2011
|
|
5BU3
 
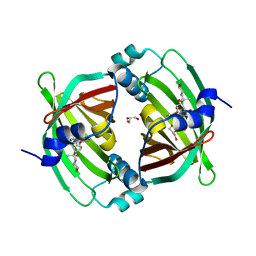 | | Crystal Structure of Diels-Alderase PyrI4 in complex with its product | | Descriptor: | (4S,4aS,6aS,8R,9R,10aR,13R,14aS,18aR,18bR)-9-ethyl-4,8,19-trihydroxy-10a,12,13,18a-tetramethyl-2,3,4,4a,5,6,6a,7,8,9,10,10a,13,14,18a,18b-hexadecahydro-1H-14a,17-(metheno)benzo[b]naphtho[2,1-h]azacyclododecine-16,18(15H,17H)-dione, GLYCEROL, PyrI4 | | Authors: | Pan, L, Guo, Y, Liu, J. | | Deposit date: | 2015-06-03 | | Release date: | 2016-02-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.897 Å) | | Cite: | Enzyme-Dependent [4 + 2] Cycloaddition Depends on Lid-like Interaction of the N-Terminal Sequence with the Catalytic Core in PyrI4
Cell Chem Biol, 23, 2016
|
|
1EQ7
 
 | |
1RZW
 
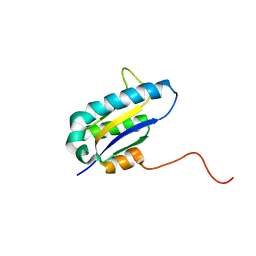 | | The Solution Structure of the Archaeglobus fulgidis protein AF2095. Northeast Structural Genomics Consortium target GR4 | | Descriptor: | Protein AF2095(GR4) | | Authors: | Powers, R, Acton, T.B, Huang, Y.J, Liu, J, Ma, L, Rost, B, Chiang, Y, Cort, J.R, Kennedy, M.A, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-12-29 | | Release date: | 2004-11-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Archaeglobus fulgidis peptidyl-tRNA hydrolase (Pth2) provides evidence for an extensive conserved family of Pth2 enzymes in archea, bacteria, and eukaryotes
Protein Sci., 14, 2005
|
|
8DSP
 
 | |
