7OTF
 
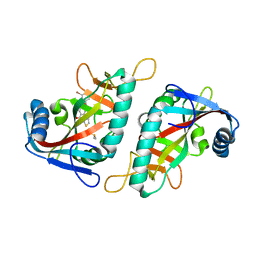 | | PARP15 catalytic domain in complex with OUL213 | | Descriptor: | 8-[(3-bromophenyl)methoxy]-4~{H}-thieno[2,3-c]isoquinolin-5-one, DIMETHYL SULFOXIDE, Protein mono-ADP-ribosyltransferase PARP15 | | Authors: | Maksimainen, M.M, Lehtio, L. | | Deposit date: | 2021-06-10 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Analogs of TIQ-A as inhibitors of human mono-ADP-ribosylating PARPs.
Bioorg.Med.Chem., 52, 2021
|
|
7OQQ
 
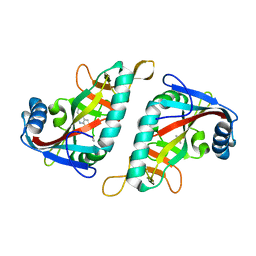 | | PARP15 catalytic domain in complex with TIQ-A | | Descriptor: | 4H-thieno[2,3-c]isoquinolin-5-one, DIMETHYL SULFOXIDE, Protein mono-ADP-ribosyltransferase PARP15 | | Authors: | Maksimainen, M.M, Lehtio, L. | | Deposit date: | 2021-06-04 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Analogs of TIQ-A as inhibitors of human mono-ADP-ribosylating PARPs.
Bioorg.Med.Chem., 52, 2021
|
|
7OSM
 
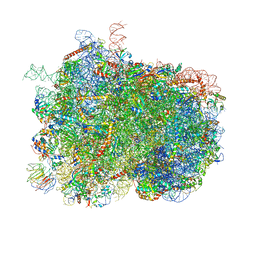 | | Intermediate translocation complex of 80 S.cerevisiae ribosome with eEF2 and ligands | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0, ... | | Authors: | Djumagulov, M, Jenner, L, Rozov, A, Demeshkina, N, Yusupov, M, Yusupova, G. | | Deposit date: | 2021-06-09 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Accuracy mechanism of eukaryotic ribosome translocation.
Nature, 600, 2021
|
|
8TUC
 
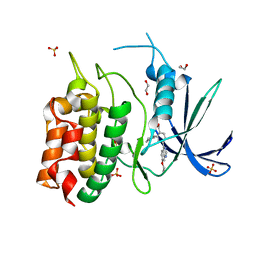 | | Unphosphorylated CaMKK2 in complex with CC-8977 | | Descriptor: | (4M)-2-cyclopentyl-4-(7-ethoxyquinazolin-4-yl)benzoic acid, 1,2-ETHANEDIOL, Calcium/calmodulin-dependent protein kinase kinase 2, ... | | Authors: | Bernard, S.M, Shanmugasundaram, V, D'Agostino, L. | | Deposit date: | 2023-08-16 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Identification of Small Molecule Inhibitors and Ligand Directed Degraders of Calcium/Calmodulin Dependent Protein Kinase Kinase 1 and 2 (CaMKK1/2).
J.Med.Chem., 66, 2023
|
|
8Z9Z
 
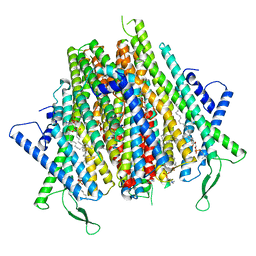 | | Cryo-EM structure of the insect olfactory receptor OR5-Orco heterocomplex from Acyrthosiphon pisum | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Odorant receptor, ApisOR5, ... | | Authors: | Wang, Y.D, Qiu, L, Guan, Z.Y, Wang, Q, Wang, G.R, Yin, P. | | Deposit date: | 2024-04-24 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for odorant recognition of the insect odorant receptor OR-Orco heterocomplex.
Science, 384, 2024
|
|
8Z9A
 
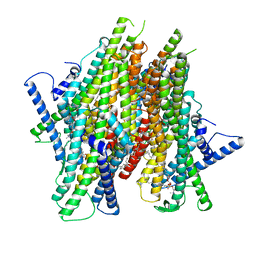 | | Cryo-EM structure of the insect olfactory receptor OR5-Orco heterocomplex from Acyrthosiphon pisum bound with geranyl acetate | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Odorant receptor, ApisOR5, ... | | Authors: | Wang, Y.D, Qiu, L, Guan, Z.Y, Wang, Q, Wang, G.R, Yin, P. | | Deposit date: | 2024-04-23 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for odorant recognition of the insect odorant receptor OR-Orco heterocomplex.
Science, 384, 2024
|
|
8TV1
 
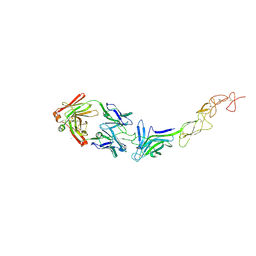 | | Structure of the EphA2 LBDCRD bound to FabS1C_L1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Ephrin type-A receptor 2, ... | | Authors: | Singer, A.U, Bruce, H.A, Blazer, L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-08-17 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Synthetic Antibodies targeting EPHA2 Induce Diverse Signaling-Competent Clusters with Differential Activation
To be published
|
|
8UAJ
 
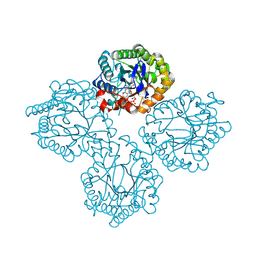 | | Succinate Bound Crystal Structure of Thermus scotoductus SA-01 Ene-reductase | | Descriptor: | FLAVIN MONONUCLEOTIDE, NADPH dehydrogenase, SUCCINIC ACID | | Authors: | Wilson, L.A, Guddat, L, Schenk, G, Scott, C. | | Deposit date: | 2023-09-21 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structural Characterization of Enzymatic Interactions with Functional Nicotinamide Cofactor Biomimetics
Catalysts, 14, 2024
|
|
8TRV
 
 | | Structure of the EphA2 LBDCRD bound to FabS1C_C1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Ephrin type-A receptor 2, ... | | Authors: | Singer, A.U, Bruce, H.A, Blazer, L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-08-10 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Synthetic Antibodies targeting EPHA2 Induce Diverse Signaling-Competent Clusters with Differential Activation
To be Published
|
|
8TV2
 
 | | Structure of apo FabS1C_L1 | | Descriptor: | CHLORIDE ION, S1C variant of Fab_L1 heavy chain, S1C variant of Fab_L1 light chain, ... | | Authors: | Singer, A.U, Bruce, H.A, Blazer, L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-08-17 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Synthetic Antibodies targeting EPHA2 Induce Diverse Signaling-Competent Clusters with Differential Activation
To be published
|
|
8TV5
 
 | | Structure of the EphA2 LBDCRD bound to FabS1CE_L1 in a 2:1 (EphA2 to Fab) ratio | | Descriptor: | Ephrin type-A receptor 2, MAGNESIUM ION, S1CE variant of Fab_L1 heavy chain, ... | | Authors: | Singer, A.U, Bruce, H.A, Blazer, L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-08-17 | | Release date: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (4.6 Å) | | Cite: | Synthetic Antibodies targeting EPHA2 Induce Diverse Signaling-Competent Clusters with Differential Activation
To be published
|
|
8UYS
 
 | | SARS-CoV-2 5' proximal stem-loop 5 | | Descriptor: | SARS-CoV-2 RNA SL5 domain. | | Authors: | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | Deposit date: | 2023-11-14 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UYJ
 
 | | BtCoV-HKU5 5' proximal stem-loop 5, conformation 4 | | Descriptor: | BtCoV-HKU5 5' proximal stem-loop 5, conformation 4 | | Authors: | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | Deposit date: | 2023-11-13 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UYL
 
 | | MERS 5' proximal stem-loop 5, conformation 2 | | Descriptor: | MERS 5' proximal stem-loop 5 | | Authors: | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | Deposit date: | 2023-11-13 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UYK
 
 | | MERS 5' proximal stem-loop 5, conformation 1 | | Descriptor: | MERS 5' proximal stem-loop 5 | | Authors: | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | Deposit date: | 2023-11-13 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UYM
 
 | | MERS 5' proximal stem-loop 5, conformation 3 | | Descriptor: | MERS 5' proximal stem-loop 5 | | Authors: | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | Deposit date: | 2023-11-13 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UYG
 
 | | BtCoV-HKU5 5' proximal stem-loop 5, conformation 2 | | Descriptor: | RNA (135-MER) | | Authors: | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | Deposit date: | 2023-11-13 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UYE
 
 | | BtCoV-HKU5 5' proximal stem-loop 5, conformation 1 | | Descriptor: | BtCoV-HKU5 5' proximal stem-loop 5 | | Authors: | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | Deposit date: | 2023-11-13 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UNH
 
 | | Cryo-EM structure of T4 Bacteriophage Clamp Loader with Sliding Clamp | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Sliding clamp, ... | | Authors: | Huang, Y, Marcus, K, Subramanian, S, Gee, L.C, Gorday, K, Ghaffari-Kashani, S, Luo, X, Zhang, L, O'Donnell, M, Subramanian, S, Kuriyan, J. | | Deposit date: | 2023-10-19 | | Release date: | 2023-12-13 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Autoinhibition of a clamp-loader ATPase revealed by deep mutagenesis and cryo-EM.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8UNF
 
 | | Cryo-EM structure of T4 Bacteriophage Clamp Loader with Sliding Clamp and DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Sliding clamp, ... | | Authors: | Huang, Y, Marcus, K, Subramanian, S, Gee, L.C, Gorday, K, Ghaffari-Kashani, S, Luo, X, Zhang, L, O'Donnell, M, Subramanian, S, Kuriyan, J. | | Deposit date: | 2023-10-18 | | Release date: | 2023-12-13 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Autoinhibition of a clamp-loader ATPase revealed by deep mutagenesis and cryo-EM.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8UYP
 
 | | SARS-CoV-1 5' proximal stem-loop 5 | | Descriptor: | SARS-CoV-1 5' proximal stem-loop 5 | | Authors: | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | Deposit date: | 2023-11-13 | | Release date: | 2023-12-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
11BG
 
 | | A POTENTIAL ALLOSTERIC SUBSITE GENERATED BY DOMAIN SWAPPING IN BOVINE SEMINAL RIBONUCLEASE | | Descriptor: | PROTEIN (BOVINE SEMINAL RIBONUCLEASE), SULFATE ION, URIDYLYL-2'-5'-PHOSPHO-GUANOSINE | | Authors: | Vitagliano, L, Adinolfi, S, Sica, F, Merlino, A, Zagari, A, Mazzarella, L. | | Deposit date: | 1999-03-11 | | Release date: | 1999-11-05 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A potential allosteric subsite generated by domain swapping in bovine seminal ribonuclease.
J.Mol.Biol., 293, 1999
|
|
7PFP
 
 | | Full-length cryo-EM structure of the native human uromodulin (UMOD)/Tamm-Horsfall protein (THP) filament | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Uromodulin, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jovine, L, Xu, C, Stsiapanava, A, Carroni, M, Tunyasuvunakool, K, Jumper, J, Wu, B. | | Deposit date: | 2021-08-11 | | Release date: | 2022-03-16 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structure of the decoy module of human glycoprotein 2 and uromodulin and its interaction with bacterial adhesin FimH.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6U91
 
 | | Crystal structure of DNMT3B(Q772R)-DNMT3L in complex with CpGpT DNA | | Descriptor: | CpGpT DNA (25-MER), DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3B, ... | | Authors: | Gao, L, Song, J. | | Deposit date: | 2019-09-06 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.99998879 Å) | | Cite: | Comprehensive structure-function characterization of DNMT3B and DNMT3A reveals distinctive de novo DNA methylation mechanisms
To Be Published
|
|
6UKO
 
 | | Structure analysis of full-length mouse bcs1 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Mitochondrial chaperone BCS1 | | Authors: | Xia, D, Esser, L. | | Deposit date: | 2019-10-05 | | Release date: | 2020-02-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | Structures of AAA protein translocase Bcs1 suggest translocation mechanism of a folded protein.
Nat.Struct.Mol.Biol., 27, 2020
|
|
