3JCP
 
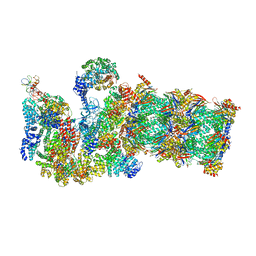 | | Structure of yeast 26S proteasome in M2 state derived from Titan dataset | | Descriptor: | 26S protease regulatory subunit 4 homolog, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B homolog, ... | | Authors: | Luan, B, Huang, X.L, Wu, J.P, Shi, Y.G, Wang, F. | | Deposit date: | 2016-01-06 | | Release date: | 2016-06-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structure of an endogenous yeast 26S proteasome reveals two major conformational states.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1U4S
 
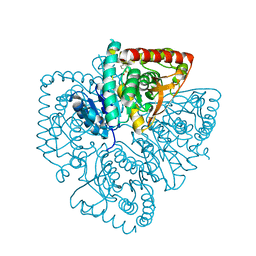 | | Plasmodium falciparum lactate dehydrogenase complexed with 2,6-naphthalenedisulphonic acid | | Descriptor: | L-lactate dehydrogenase, NAPHTHALENE-2,6-DISULFONIC ACID | | Authors: | Conners, R, Cameron, A, Read, J, Schambach, F, Sessions, R.B, Brady, R.L. | | Deposit date: | 2004-07-26 | | Release date: | 2005-06-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mapping the binding site for gossypol-like inhibitors of Plasmodium falciparum lactate dehydrogenase.
Mol.Biochem.Parasitol., 142, 2005
|
|
4V1L
 
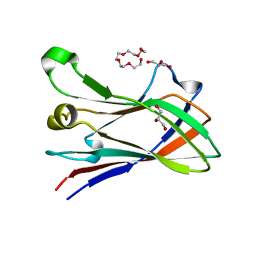 | | High resolution structure of a novel carbohydrate binding module from glycoside hydrolase family 9 (Cel9A) from Ruminococcus flavefaciens FD-1 | | Descriptor: | CARBOHYDRATE BINDING MODULE, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | Authors: | Venditto, I, Goyal, A, Thompson, A, Ferreira, L.M.A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2014-09-29 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Complexity of the Ruminococcus Flavefaciens Cellulosome Reflects an Expansion in Glycan Recognition.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4UUM
 
 | |
6XKK
 
 | | Cryo-EM structure of the NLRP1-CARD filament | | Descriptor: | NACHT, LRR and PYD domains-containing protein 1 | | Authors: | Hollingsworth, L.R, David, L, Li, Y, Sharif, H, Fontana, P, Fu, T, Wu, H. | | Deposit date: | 2020-06-26 | | Release date: | 2020-11-25 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Mechanism of filament formation in UPA-promoted CARD8 and NLRP1 inflammasomes.
Nat Commun, 12, 2021
|
|
3J22
 
 | | The Enterovirus 71 A-particle | | Descriptor: | capsid protein VP0, capsid protein VP1, capsid protein VP3 | | Authors: | Shingler, K.L. | | Deposit date: | 2012-08-13 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | The Enterovirus 71 A-particle Forms a Gateway to Allow Genome Release: A CryoEM Study of Picornavirus Uncoating.
Plos Pathog., 9, 2013
|
|
4UVR
 
 | | Binding mode, selectivity and potency of N-indolyl-oxopyridinyl-4- amino-propanyl-based inhibitors targeting Trypanosoma cruzi CYP51 | | Descriptor: | Nalpha-{4-[4-(5-chloro-2-methylphenyl)piperazin-1-yl]-2-fluorobenzoyl}-N-pyridin-4-yl-D-tryptophanamide, PROTOPORPHYRIN IX CONTAINING FE, STEROL 14-DEMETHYLASE, ... | | Authors: | Vieira, D.F, Choi, J.Y, Calvet, C.M, Gut, J, Kellar, D, Siqueira-Neto, J.L, Johnston, J.B, McKerrow, J.H, Roush, W.R, Podust, L.M. | | Deposit date: | 2014-08-08 | | Release date: | 2014-11-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Binding Mode and Potency of N-Indolyl-Oxopyridinyl-4-Amino-Propanyl-Based Inhibitors Targeting Trypanosoma Cruzi Cyp51
J.Med.Chem., 57, 2014
|
|
1U4O
 
 | | Plasmodium falciparum lactate dehydrogenase complexed with 2,6-naphthalenedicarboxylic acid | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2,6-DICARBOXYNAPHTHALENE, L-lactate dehydrogenase | | Authors: | Conners, R, Cameron, A, Read, J, Schambach, F, Sessions, R.B, Brady, R.L. | | Deposit date: | 2004-07-26 | | Release date: | 2005-06-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mapping the binding site for gossypol-like inhibitors of Plasmodium falciparum lactate dehydrogenase.
Mol.Biochem.Parasitol., 142, 2005
|
|
1U5C
 
 | | Plasmodium falciparum lactate dehydrogenase complexed with 3,7-dihydroxynaphthalene-2-carboxylic acid and NAD+ | | Descriptor: | 3,7-DIHYDROXY-2-NAPHTHOIC ACID, L-lactate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Conners, R, Cameron, A, Read, J, Schambach, F, Sessions, R.B, Brady, R.L. | | Deposit date: | 2004-07-27 | | Release date: | 2005-06-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Mapping the binding site for gossypol-like inhibitors of Plasmodium falciparum lactate dehydrogenase.
Mol.Biochem.Parasitol., 142, 2005
|
|
5XF1
 
 | | Structure of the Full-length glucagon class B G protein-coupled receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, H, Qiao, A, Yang, D, Yang, L, Dai, A, de Graaf, C, Reedtz-Runge, S, Dharmarajan, V, Zhang, H, Han, G.W, Grant, T, Sierra, R, Weierstall, U, Nelson, G, Liu, W, Wu, Y, Ma, L, Cai, X, Lin, G, Wu, X, Geng, Z, Dong, Y, Song, G, Griffin, P, Lau, J, Cherezov, V, Yang, H, Hanson, M, Stevens, R, Jiang, H, Wang, M, Zhao, Q, Wu, B. | | Deposit date: | 2017-04-06 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Structure of the full-length glucagon class B G-protein-coupled receptor.
Nature, 546, 2017
|
|
6ORR
 
 | | Co-crystal structure of human NicotinamideN-Methyltransferase (NNMT) in complex with High-Affinity Alkynyl Bisubstrate Inhibitor NS1 | | Descriptor: | 9-{9-amino-6-[(3-carbamoylphenyl)ethynyl]-5,6,7,8,9-pentadeoxy-D-glycero-alpha-L-talo-decofuranuronosyl}-9H-purin-6-amine, GLYCEROL, NNMT protein | | Authors: | May, E.J, Policarpo, R.L, Gaudet, R. | | Deposit date: | 2019-04-30 | | Release date: | 2019-10-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | High-Affinity Alkynyl Bisubstrate Inhibitors of NicotinamideN-Methyltransferase (NNMT).
J.Med.Chem., 62, 2019
|
|
1KNG
 
 | | Crystal structure of CcmG reducing oxidoreductase at 1.14 A | | Descriptor: | THIOL:DISULFIDE INTERCHANGE PROTEIN CYCY | | Authors: | Edeling, M.A, Guddat, L.W, Fabianek, R.A, Thony-Meyer, L, Martin, J.L. | | Deposit date: | 2001-12-18 | | Release date: | 2002-07-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Structure of CcmG/DsbE at 1.14 A resolution: high-fidelity reducing activity in an indiscriminately oxidizing environment
Structure, 10, 2002
|
|
1U26
 
 | | Crystal structure of Selenomonas ruminantium phytase complexed with persulfated phytate | | Descriptor: | D-MYO-INOSITOL-HEXASULPHATE, myo-inositol hexaphosphate phosphohydrolase | | Authors: | Chu, H.M, Guo, R.T, Lin, T.W, Chou, C.C, Shr, H.L, Lai, H.L, Tang, T.Y, Cheng, K.J, Selinger, B.L, Wang, A.H.-J. | | Deposit date: | 2004-07-16 | | Release date: | 2004-11-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of Selenomonas ruminantium Phytase in Complex with Persulfated Phytate; DSP Phytase Fold and Mechanism for Sequential Substrate Hydrolysis
STRUCTURE, 12, 2004
|
|
1U7H
 
 | | Structure and a Proposed Mechanism for Ornithine Cyclodeaminase from Pseudomonas putida | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Alam, S, Goodman, J.L, Wang, S, Ruzicka, F.J, Frey, P.A, Wedekind, J.E. | | Deposit date: | 2004-08-03 | | Release date: | 2004-11-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ornithine Cyclodeaminase: Structure, Mechanism of Action, and Implications for
the u-Crystallin Family;
Biochemistry, 43, 2004
|
|
5I4D
 
 | | 1.75 Angstrom Crystal Structure of Superantigen-like Protein, Exotoxin from Staphylococcus aureus, in Complex with Sialyl-LewisX. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-alpha-D-glucopyranose, Superantigen-like protein, ... | | Authors: | Minasov, G, Nocadello, S, Shuvalova, L, Filippova, E.V, Halavaty, A, Dubrovska, I, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-02-11 | | Release date: | 2016-02-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | 1.75 Angstrom Crystal Structure of Superantigen-like Protein, Exotoxin from Staphylococcus aureus, in Complex with Sialyl-LewisX.
To Be Published
|
|
5X08
 
 | | Crystal structure of broadly neutralizing anti-HIV-1 antibody 4E10, mutant Npro, with peptide bound | | Descriptor: | ACETATE ION, CHLORIDE ION, Envelope glycoprotein gp160, ... | | Authors: | Caaveiro, J.M.M, Rujas, E, Nieva, J.L, Tsumoto, K. | | Deposit date: | 2017-01-20 | | Release date: | 2017-04-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Functional Contacts between MPER and the Anti-HIV-1 Broadly Neutralizing Antibody 4E10 Extend into the Core of the Membrane
J. Mol. Biol., 429, 2017
|
|
8IJC
 
 | | NMR solution structure of the 1:1 complex of a platinum(II) ligand L1-transpt covalently bound to a G-quadruplex MYT1L | | Descriptor: | G-quadruplex DNA MYT1L, Pt(NH3)2(2-(pyridin-4-ylmethyl)benzo-[lmn][3,8]phenanthroline-1,3,6,8(2H,7H)-tetraone) | | Authors: | Liu, L.-Y, Mao, Z.-W. | | Deposit date: | 2023-02-27 | | Release date: | 2023-06-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Organic-Platinum Hybrids for Covalent Binding of G-Quadruplexes: Structural Basis and Application to Cancer Immunotherapy.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
5XS5
 
 | | Structure of Coxsackievirus A6 (CVA6) virus procapsid particle | | Descriptor: | Genome polyprotein | | Authors: | Zheng, Q.B, He, M.Z, Xu, L.F, Yu, H, Cheng, T, Li, S.W. | | Deposit date: | 2017-06-12 | | Release date: | 2017-09-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Atomic structures of Coxsackievirus A6 and its complex with a neutralizing antibody
Nat Commun, 8, 2017
|
|
7N6I
 
 | | ATP-bound TnsC-TniQ complex from ShCAST system | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Park, J, Tsai, A.W.L, Mehrotra, E, Kellogg, E.H. | | Deposit date: | 2021-06-08 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for target site selection in RNA-guided DNA transposition systems.
Science, 373, 2021
|
|
6OF7
 
 | | Crystal structure of the CRY1-PER2 complex | | Descriptor: | Cryptochrome-1, Period circadian protein homolog 2 | | Authors: | Michael, A.K, Fribourgh, J.L, Tripathi, S.M, Partch, C.L. | | Deposit date: | 2019-03-28 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Dynamics at the serine loop underlie differential affinity of cryptochromes for CLOCK:BMAL1 to control circadian timing.
Elife, 9, 2020
|
|
2OZ4
 
 | | Structural Plasticity in IgSF Domain 4 of ICAM-1 Mediates Cell Surface Dimerization | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, FAB FRAGMENT LIGHT CHAIN, ... | | Authors: | Chen, X, Kim, T.D, Carman, C.V, Mi, L, Song, G, Springer, T.A. | | Deposit date: | 2007-02-23 | | Release date: | 2007-10-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural plasticity in Ig superfamily domain 4 of ICAM-1 mediates cell surface dimerization.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
8IF7
 
 | | Crystal structure of CmnB | | Descriptor: | 2-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]ACRYLIC ACID, CmnB | | Authors: | Chang, C.Y, Toh, S.I, Lo, C.L. | | Deposit date: | 2023-02-17 | | Release date: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of CmnB involved in the biosynthesis of the nonproteinogenic amino acid L-2,3-diaminopropionic acid.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
5XM6
 
 | | the overall structure of VrEH2 | | Descriptor: | Epoxide hydrolase | | Authors: | Li, F.L, Kong, X.D, Yu, H.L, Shang, Y.P, Zhou, J.H, Xu, J.H. | | Deposit date: | 2017-05-12 | | Release date: | 2018-09-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Regioselectivity Engineering of Epoxide Hydrolase: Near-Perfect Enantioconvergence through a Single Site Mutation
Acs Catalysis, 8, 2018
|
|
6OLL
 
 | |
6ORV
 
 | | Non-peptide agonist (TT-OAD2) bound to the Glucagon-Like peptide-1 (GLP-1) Receptor | | Descriptor: | Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Belousoff, M.J, Liang, Y.L, Danev, R. | | Deposit date: | 2019-05-01 | | Release date: | 2020-01-08 | | Last modified: | 2020-01-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Activation of the GLP-1 receptor by a non-peptidic agonist.
Nature, 577, 2020
|
|
