1V0A
 
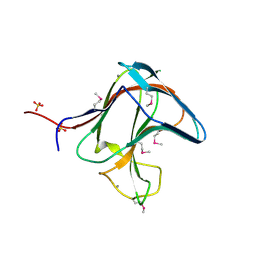 | | Family 11 Carbohydrate-Binding Module of cellulosomal cellulase Lic26A-Cel5E of Clostridium thermocellum | | Descriptor: | CALCIUM ION, ENDOGLUCANASE H, SULFATE ION | | Authors: | Carvalho, A.L, Romao, M.J, Goyal, A, Prates, J.A.M, Pires, V.M.R, Ferreira, L.M.A, Bolam, D.N, Gilbert, H.J, Fontes, C.M.G.A. | | Deposit date: | 2004-03-25 | | Release date: | 2005-01-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The Family 11 Carbohydrate-Binding Module of Clostridium Thermocellum Lic26A-Cel5E Accomodates Beta-1,4- and Beta-1,3-1,4-Mixed Linked Glucans at a Single Binding Site
J.Biol.Chem., 279, 2004
|
|
3TEO
 
 | | APO Form of carbon disulfide hydrolase (selenomethionine form) | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, CHLORIDE ION, Carbon disulfide hydrolase | | Authors: | Smeulders, M.J, Barends, T.R.M.B, Pol, A, Scherer, A, Zandvoort, M.H, Udvarhelyi, A, Khadem, A, Menzel, A, Hermans, J, Shoeman, R.L, Wessels, H.J.C.T, van den Heuvel, L.P, Russ, L, Schlichting, I, Jetten, M.S.M, Op den Camp, H.J.M. | | Deposit date: | 2011-08-15 | | Release date: | 2011-10-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Evolution of a new enzyme for carbon disulphide conversion by an acidothermophilic archaeon.
Nature, 478, 2011
|
|
2F3Y
 
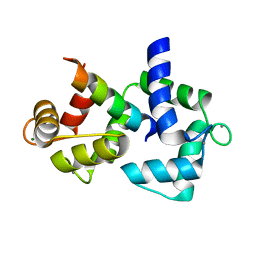 | | Calmodulin/IQ domain complex | | Descriptor: | CALCIUM ION, Calmodulin, MAGNESIUM ION, ... | | Authors: | Fallon, J.L, Quiocho, F.A. | | Deposit date: | 2005-11-22 | | Release date: | 2005-12-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of Calmodulin Bound to the Hydrophobic IQ Domain of the Cardiac Ca(v)1.2 Calcium Channel.
Structure, 13, 2005
|
|
5XEZ
 
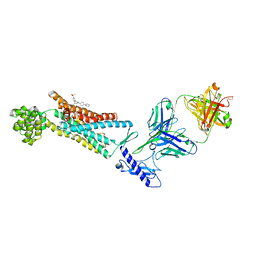 | | Structure of the Full-length glucagon class B G protein-coupled receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-{[(4-cyclohexylphenyl){[3-(methylsulfonyl)phenyl]carbamoyl}amino]methyl}-N-(1H-tetrazol-5-yl)benzamide, ... | | Authors: | Zhang, H, Qiao, A, Yang, D, Yang, L, Dai, A, de Graaf, C, Reedtz-Runge, S, Dharmarajan, V, Zhang, H, Han, G.W, Grant, T, Sierra, R, Weierstall, U, Nelson, G, Liu, W, Wu, Y, Ma, L, Cai, X, Lin, G, Wu, X, Geng, Z, Dong, Y, Song, G, Griffin, P, Lau, J, Cherezov, V, Yang, H, Hanson, M, Stevens, R, Jiang, H, Wang, M, Zhao, Q, Wu, B. | | Deposit date: | 2017-04-06 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the full-length glucagon class B G-protein-coupled receptor.
Nature, 546, 2017
|
|
1L3T
 
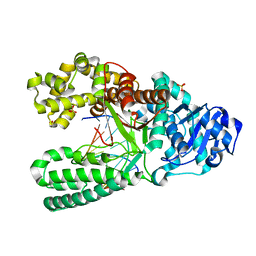 | | Crystal Structure of Bacillus DNA Polymerase I Fragment product complex with 10 base pairs of duplex DNA following addition of a single dTTP residue | | Descriptor: | 5'-D(*GP*AP*CP*G*TP*AP*CP*GP*TP*GP*AP*TP*CP*GP*CP*A)-3', 5'-D(*GP*CP*GP*AP*TP*CP*AP*CP*GP*T)-3', DNA Polymerase I, ... | | Authors: | Johnson, S.J, Taylor, J.S, Beese, L.S. | | Deposit date: | 2002-03-01 | | Release date: | 2003-03-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Processive DNA synthesis observed in a polymerase crystal suggests a
mechanism for the prevention of frameshift mutations
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
5XS7
 
 | | Structure of Coxsackievirus A6 (CVA6) virus A-particle in complex with the neutralizing antibody fragment 1D5 | | Descriptor: | Genome polyprotein, Heavy chain of Fab 1D5, Light chain of Fab 1D5 | | Authors: | Zheng, Q.B, He, M.Z, Xu, L.F, Yu, H, Li, S.W, Cheng, T. | | Deposit date: | 2017-06-12 | | Release date: | 2017-09-27 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Atomic structures of Coxsackievirus A6 and its complex with a neutralizing antibody
Nat Commun, 8, 2017
|
|
1VYZ
 
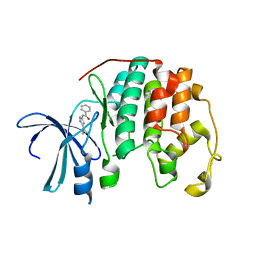 | | Structure of CDK2 complexed with PNU-181227 | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, N-(5-CYCLOPROPYL-1H-PYRAZOL-3-YL)BENZAMIDE | | Authors: | Pevarello, P, Brasca, M.G, Amici, R, Orsini, P, Traquandi, G, Corti, L, Piutti, C, Sansonna, P, Villa, M, Pierce, B.S, Pulici, M, Giordano, G, Martina, K, Lfritzen, E, Nugent, R.A, Casale, E, Cameron, A, Ciomei, M, Roletto, F, Isacchi, A, Fogliatto, G, Pesenti, E, Pastori, W, Marsiglio, W, Leach, K.L, Clare, P.M, Fiorentini, F, Varasi, M, Vulpetti, A, Warpehoski, M.A. | | Deposit date: | 2004-05-07 | | Release date: | 2004-06-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | 3-Aminopyrazole Inhibitors of Cdk2/Cyclin a as Antitumor Agents. 1. Lead Finding
J.Med.Chem., 47, 2004
|
|
4MPS
 
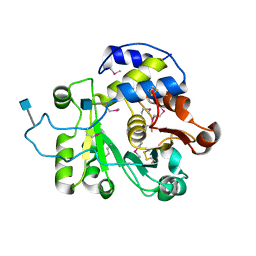 | | Crystal structure of rat Beta-galactoside alpha-2,6-sialyltransferase 1 (ST6GAL1), Northeast Structural Genomics Consortium Target RnR367A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-galactoside alpha-2,6-sialyltransferase 1 | | Authors: | Forouhar, F, Meng, L, Milaninia, S, Seetharaman, J, Su, M, Kornhaber, G, Montelione, G.T, Hunt, J.F, Moremen, K.W, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-09-13 | | Release date: | 2013-09-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Enzymatic Basis for N-Glycan Sialylation: STRUCTURE OF RAT alpha 2,6-SIALYLTRANSFERASE (ST6GAL1) REVEALS CONSERVED AND UNIQUE FEATURES FOR GLYCAN SIALYLATION.
J.Biol.Chem., 288, 2013
|
|
5J3Z
 
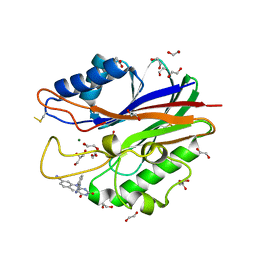 | | Crystal structure of m2hTDP2-CAT in complex with a small molecule inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2,4-dioxo-10-[3-(1H-tetrazol-5-yl)phenyl]-2,3,4,10-tetrahydropyrimido[4,5-b]quinoline-8-carbonitrile, ACETATE ION, ... | | Authors: | Hornyak, P, Pearl, L.H, Caldecott, K.W, Oliver, A.W. | | Deposit date: | 2016-03-31 | | Release date: | 2016-05-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mode of action of DNA-competitive small molecule inhibitors of tyrosyl DNA phosphodiesterase 2.
Biochem.J., 473, 2016
|
|
8BG1
 
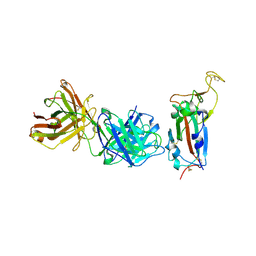 | | Crystal structure of the SARS-CoV-2 S RBD in complex with pT1511 scFV | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hansen, G, Ssebyatika, G.L, Krey, T. | | Deposit date: | 2022-10-27 | | Release date: | 2023-11-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Activity of broadly neutralizing antibodies against sarbecoviruses: a trade-off between SARS-CoV-2 variants and distant coronaviruses?
To be published
|
|
1VKC
 
 | | Putative acetyl transferase from Pyrococcus furiosus | | Descriptor: | IODIDE ION, putative acetyl transferase | | Authors: | Habel, J.E, Liu, Z.-J, Tempel, W, Rose, J.P, Brereton, P.S, Izumi, M, Jenney Jr, F.E, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Richardson, D.C, Richardson, J.S, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-05-11 | | Release date: | 2004-12-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Putative acetyl transferase from Pyrococcus furiosus
To be published
|
|
1LQO
 
 | | Crystal Strutcure of the Fosfomycin Resistance Protein A (FosA) Containing Bound Thallium Cations | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, PROBABLE Fosfomycin Resistance Protein, ... | | Authors: | Rife, C.L, Pharris, R.E, Newcomer, M.E, Armstrong, R.N. | | Deposit date: | 2002-05-11 | | Release date: | 2002-09-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a genomically encoded fosfomycin resistance protein (FosA) at 1.19 A resolution by MAD
phasing off the L-III edge of Tl(+)
J.Am.Chem.Soc., 124, 2002
|
|
8NSE
 
 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE, NNA COMPLEX | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, CACODYLIC ACID, GLYCEROL, ... | | Authors: | Raman, C.S, Li, H, Martasek, P, Masters, B.S.S, Poulos, T.L. | | Deposit date: | 1999-01-14 | | Release date: | 2001-11-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of nitric oxide synthase bound to nitro indazole reveals a novel inactivation mechanism.
Biochemistry, 40, 2001
|
|
4N9F
 
 | | Crystal structure of the Vif-CBFbeta-CUL5-ElOB-ElOC pentameric complex | | Descriptor: | Core-binding factor subunit beta, Cullin-5, Transcription elongation factor B polypeptide 1, ... | | Authors: | Guo, Y.Y, Dong, L.Y, Huang, Z.W. | | Deposit date: | 2013-10-21 | | Release date: | 2014-01-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for hijacking CBF-b and CUL5 E3 ligase complex by HIV-1 Vif
Nature, 505, 2014
|
|
4EEV
 
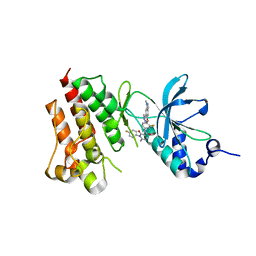 | | Crystal structure of c-Met in complex with LY2801653 | | Descriptor: | Hepatocyte growth factor receptor, N-(3-fluoro-4-{[1-methyl-6-(1H-pyrazol-4-yl)-1H-indazol-5-yl]oxy}phenyl)-1-(4-fluorophenyl)-6-methyl-2-oxo-1,2-dihydropyridine-3-carboxamide | | Authors: | Wang, Y, Stout, S.L. | | Deposit date: | 2012-03-28 | | Release date: | 2013-04-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | LY2801653 is an orally bioavailable multi-kinase inhibitor with potent activity against MET, MST1R, and other oncoproteins, and displays anti-tumor activities in mouse xenograft models.
Invest New Drugs, 31, 2013
|
|
1LQP
 
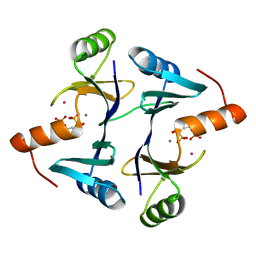 | | CRYSTAL STRUCTURE OF THE FOSFOMYCIN RESISTANCE PROTEIN (FOSA) CONTAINING BOUND SUBSTRATE | | Descriptor: | FOSFOMYCIN, MANGANESE (II) ION, POTASSIUM ION, ... | | Authors: | Rife, C.L, Pharris, R.E, Newcomer, M.E, Armstrong, R.N. | | Deposit date: | 2002-05-11 | | Release date: | 2002-09-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Crystal structure of a genomically encoded fosfomycin resistance protein (FosA) at 1.19 A resolution by MAD
phasing off the L-III edge of Tl(+)
J.Am.Chem.Soc., 124, 2002
|
|
1W93
 
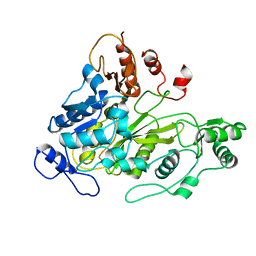 | | Crystal Structure of Biotin Carboxylase Domain of Acetyl-Coenzyme A Carboxylase from Saccharomyces cerevisiae | | Descriptor: | ACETYL-COENZYME A CARBOXYLASE | | Authors: | Shen, Y, Volrath, S.L, Weatherly, S.C, Elich, T.D, Tong, L. | | Deposit date: | 2004-10-05 | | Release date: | 2005-01-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Mechanism for the Potent Inhibition of Eukaryotic Acetyl-Coenzyme a Carboxylase by Soraphen A, a Macrocyclic Polyketide Natural Product
Mol.Cell, 16, 2004
|
|
1LRT
 
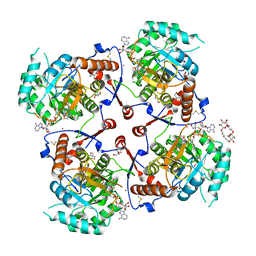 | | CRYSTAL STRUCTURE OF TERNARY COMPLEX OF TRITRICHOMONAS FOETUS INOSINE-5'-MONOPHOSPHATE DEHYDROGENASE: STRUCTURAL CHARACTERIZATION OF NAD+ SITE IN MICROBIAL ENZYME | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-METHYLENE-THIAZOLE-4-CARBOXYAMIDE-ADENINE DINUCLEOTIDE, INOSINE-5'-MONOPHOSPHATE DEHYDROGENASE, ... | | Authors: | Gan, L, Petsko, G.A, Hedstrom, L. | | Deposit date: | 2002-05-15 | | Release date: | 2003-07-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a ternary complex of Tritrichomonas foetus inosine
5'-monophosphate dehydrogenase: NAD+ orients the active site loop for catalysis
Biochemistry, 41, 2003
|
|
5XU9
 
 | | Crystal Structure of Transketolase in complex with TPP intermediate IX and gauche form erythrose-4-phosphate from Pichia Stipitis | | Descriptor: | 2-[(2E)-3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-2-[1,2-bis(oxidanyl)ethylidene]-4-methyl-1,3-thiazol-5-yl]ethyl phosphono hydrogen phosphate, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Li, T.L, Hsu, N.S, Wang, Y.L. | | Deposit date: | 2017-06-22 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.166 Å) | | Cite: | Evidence of Diradicals Involved in the Yeast Transketolase Catalyzed Keto-Transferring Reactions.
Chembiochem, 19, 2018
|
|
8OIU
 
 | | Cryo-EM reconstruction of the native 24-mer E2o core of the 2-oxoglutarate dehydrogenase complex of C. thermophilum at 3.35 A resolution | | Descriptor: | Dihydrolipoyllysine-residue succinyltransferase | | Authors: | Skalidis, I, Tueting, C, Kyrilis, F.L, Hamdi, F, Kastritis, P.L. | | Deposit date: | 2023-03-23 | | Release date: | 2023-05-31 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural analysis of an endogenous 4-megadalton succinyl-CoA-generating metabolon.
Commun Biol, 6, 2023
|
|
3BRT
 
 | | NEMO/IKK association domain structure | | Descriptor: | Inhibitor of nuclear factor kappa-B kinase subunit beta,Inhibitor of nuclear factor kappa-B kinase subunit alpha, NF-kappa-B essential modulator | | Authors: | Silvian, L.F. | | Deposit date: | 2007-12-21 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of a NEMO/IKK-Associating Domain Reveals Architecture of the Interaction Site.
Structure, 16, 2008
|
|
5JK2
 
 | | Crystal structure of Treponema pallidum Tp0751 (Pallilysin) | | Descriptor: | Tp0751 | | Authors: | Parker, M.L, Boulanger, M.J. | | Deposit date: | 2016-04-25 | | Release date: | 2016-10-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Structure of Treponema pallidum Tp0751 (Pallilysin) Reveals a Non-canonical Lipocalin Fold That Mediates Adhesion to Extracellular Matrix Components and Interactions with Host Cells.
Plos Pathog., 12, 2016
|
|
3BUE
 
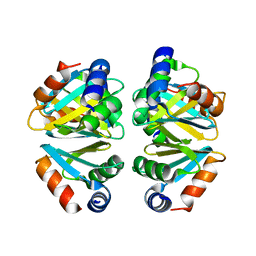 | | Crystal structure of the C-terminal domain hexamer of ArgR from Mycobacterium tuberculosis | | Descriptor: | Arginine repressor ArgR | | Authors: | Cherney, L.T, Cherney, M.M, Garen, C.R, Lu, G.J, James, M.N.G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2008-01-02 | | Release date: | 2008-01-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of the C-terminal domain of the arginine repressor protein from Mycobacterium tuberculosis.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
1W0U
 
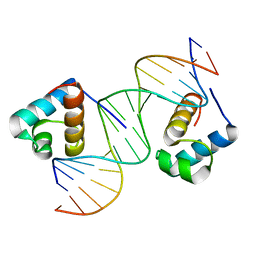 | | hTRF2 DNA-binding domain in complex with telomeric DNA. | | Descriptor: | 5'-D(*CP*TP*AP*AP*CP*CP*CP*TP*AP*AP *CP*CP*CP*TP*AP*GP*A)-3', 5'-D(*TP*CP*TP*AP*GP*GP*GP*TP*TP*AP *GP*GP*GP*TP*TP*AP*G)-3', TELOMERIC REPEAT BINDING FACTOR 2 | | Authors: | Court, R.I, Chapman, L.M, Fairall, L, Rhodes, D. | | Deposit date: | 2004-06-11 | | Release date: | 2004-12-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | How the Human Telomeric Proteins Trf1 and Trf2 Recognize Telomeric DNA: A View from High-Resolution Crystal Structures
Embo Rep., 6, 2005
|
|
5JHG
 
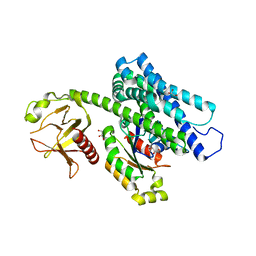 | | Crystal structure of the complex between the human RhoA and the DH/PH domain of human ARHGEF11 | | Descriptor: | GLYCEROL, Rho guanine nucleotide exchange factor 11, Transforming protein RhoA | | Authors: | Wang, R, Chen, Q, Zhang, H, Yan, Z, Li, J, Miao, L, Wang, F. | | Deposit date: | 2016-04-21 | | Release date: | 2017-04-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallization and preliminary X-ray crystallographic analysis of a small GTPase RhoA bound with its inhibitor and ARHGEF11
To Be Published
|
|
