3O9N
 
 | | Crystal Structure of a new form of xylanase-A-amylase inhibitor protein(XAIP-III) at 2.4 A resolution | | Descriptor: | ACETATE ION, Haementhin, PHOSPHATE ION | | Authors: | Singh, A, Kumar, S, Sinha, M, Sharma, S, Singh, T.P. | | Deposit date: | 2010-08-04 | | Release date: | 2010-09-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of a new form of xylanase-A-amylase inhibitor protein(XAIP-III) at 2.4 A resolution
To be Published
|
|
4EIX
 
 | | Structural Studies of the ternary complex of Phaspholipase A2 with nimesulide and indomethacin | | Descriptor: | 4-NITRO-2-PHENOXYMETHANESULFONANILIDE, ACETONITRILE, INDOMETHACIN, ... | | Authors: | Shukla, P.K, Singh, N, Kumar, S, Bhushan, A, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-04-06 | | Release date: | 2012-04-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Studies of the ternary complex of Phaspholipase A2 with nimusulide and indomethacin
To be Published
|
|
2QJE
 
 | | Crystal structure of the complex of Bovine C-lobe with Amygdalin at 2.3A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, FE (III) ION, ... | | Authors: | Mir, R, Jain, R, Kumar, S, Sinha, M, Singh, N, Sharma, S, Kaur, P, Srinivasan, A, Bhushan, A, Singh, T.P. | | Deposit date: | 2007-07-07 | | Release date: | 2008-06-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the complex of Bovine C-lobe with Amygdalin at 2.3A resolution
To be Published
|
|
2R9J
 
 | | Ligand recognition in C-lobe: The crystal structure of the complex of lactoferrin C-lobe with nicotinamide at 2.5 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, ... | | Authors: | Mir, R, Vikram, G, Singh, N, Kumar, S, Sinha, M, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2007-09-13 | | Release date: | 2007-10-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Ligand recognition in C-lobe: The crystal structure of the complex of lactoferrin C-lobe with nicotinamide at 2.5 A resolution
To be Published
|
|
1RGY
 
 | | Citrobacter freundii GN346 Class C beta-Lactamase Complexed with Transition-State Analog of Cefotaxime | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, beta-lactamase, {[(2E)-2-(2-AMINO-1,3-THIAZOL-4-YL)-2-(METHOXYIMINO)ETHANOYL]AMINO}METHYLPHOSPHONIC ACID | | Authors: | Nukaga, M, Kumar, S, Nukaga, K, Pratt, R.F, Knox, J.R. | | Deposit date: | 2003-11-13 | | Release date: | 2004-04-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Hydrolysis of third-generation cephalosporins by class C beta-lactamases. Structures of a transition state analog of cefotoxamine in wild-type and extended spectrum enzymes.
J.Biol.Chem., 279, 2004
|
|
3BC3
 
 | | Exploring inhibitor binding at the S subsites of cathepsin L | | Descriptor: | Cathepsin L heavy and light chains, S-benzyl-N-(biphenyl-4-ylacetyl)-L-cysteinyl-N~5~-(diaminomethyl)-D-ornithyl-N-(2-phenylethyl)-L-tyrosinamide | | Authors: | Chowdhury, S.F, Joseph, L, Kumar, S, Tulsidas, S.R, Bhat, S, Ziomek, E, Nard, R.M, Sivaraman, J, Purisima, E.O. | | Deposit date: | 2007-11-12 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Exploring inhibitor binding at the S' subsites of cathepsin L
J.Med.Chem., 51, 2008
|
|
3E9K
 
 | | Crystal structure of Homo sapiens kynureninase-3-hydroxyhippuric acid inhibitor complex | | Descriptor: | 3-Hydroxyhippuric acid, Kynureninase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Lima, S, Kumar, S, Gawandi, V, Momany, C, Phillips, R.S. | | Deposit date: | 2008-08-22 | | Release date: | 2008-12-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the Homo sapiens kynureninase-3-hydroxyhippuric acid inhibitor complex: insights into the molecular basis of kynureninase substrate specificity.
J.Med.Chem., 52, 2009
|
|
2R71
 
 | | Crystal structure of the complex of bovine C-lobe with inositol at 2.1A resolution | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, ... | | Authors: | Mir, R, Jain, R, Kumar, S, Sinha, M, Singh, N, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2007-09-07 | | Release date: | 2007-10-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structure of the complex of bovine C-lobe with inositol at 2.1A resolution
To be Published
|
|
1RGZ
 
 | | Enterobacter cloacae GC1 Class C beta-Lactamase Complexed with Transition-State Analog of Cefotaxime | | Descriptor: | GLYCEROL, class C beta-lactamase, {[(2E)-2-(2-AMINO-1,3-THIAZOL-4-YL)-2-(METHOXYIMINO)ETHANOYL]AMINO}METHYLPHOSPHONIC ACID | | Authors: | Nukaga, M, Kumar, S, Nukaga, K, Pratt, R.F, Knox, J.R. | | Deposit date: | 2003-11-13 | | Release date: | 2004-04-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Hydrolysis of third-generation cephalosporins by class C beta-lactamases. Structures of a transition state analog of cefotoxamine in wild-type and extended spectrum enzymes.
J.Biol.Chem., 279, 2004
|
|
8FXT
 
 | | Escherichia coli periplasmic Glucose-Binding Protein glucose complex: Acrylodan conjugate attached at W183C | | Descriptor: | 1-[6-(dimethylamino)naphthalen-2-yl]propan-1-one, CALCIUM ION, D-galactose/methyl-galactoside binding periplasmic protein MglB, ... | | Authors: | Allert, M.J, Kumar, S, Wang, Y, Beese, L.S, Hellinga, H.W. | | Deposit date: | 2023-01-25 | | Release date: | 2023-08-30 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Chromophore carbonyl twisting in fluorescent biosensors encodes direct readout of protein conformations with multicolor switching.
Commun Chem, 6, 2023
|
|
8FXU
 
 | | Thermoanaerobacter thermosaccharolyticum periplasmic Glucose-Binding Protein glucose complex: Badan conjugate attached at F17C | | Descriptor: | 2-bromo-1-[6-(dimethylamino)naphthalen-2-yl]ethan-1-one, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Allert, M.J, Kumar, S, Beese, L.S, Hellinga, H.W. | | Deposit date: | 2023-01-25 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Chromophore carbonyl twisting in fluorescent biosensors encodes direct readout of protein conformations with multicolor switching.
Commun Chem, 6, 2023
|
|
7D95
 
 | |
7D40
 
 | |
8AGA
 
 | | Structure of p-hydroxy benzoic acid ligand bound HosA transcriptional regulator from enteropathogenic Escherichia coli O127:H6 (strain E2348/69) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, P-HYDROXYBENZOIC ACID, ... | | Authors: | Goswami, A, Kannika, B.R, Madan Kumar, S. | | Deposit date: | 2022-07-19 | | Release date: | 2023-08-02 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Horizontally acquired HosA transcription factor bound with 4-hydroxy-benzoic acid exhibits unique tug-of-water dynamics
Biorxiv, 2024
|
|
4NZC
 
 | | Crystal structure of Chitinase D from Serratia proteamaculans at 1.45 Angstrom resolution | | Descriptor: | ACETATE ION, GLYCEROL, Glycoside hydrolase family 18 | | Authors: | Madhuprakash, J, Singh, A, Kumar, S, Sinha, M, Kaur, P, Sharma, S, Podile, A.R, Singh, T.P. | | Deposit date: | 2013-12-12 | | Release date: | 2014-01-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of chitinase D from Serratia proteamaculans reveals the structural basis of its dual action of hydrolysis and transglycosylation
Int J Biochem Mol Biol, 4, 2013
|
|
4FYM
 
 | |
4N42
 
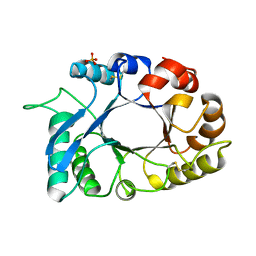 | | Crystal structure of allergen protein scam1 from Scadoxus multiflorus | | Descriptor: | PHOSPHATE ION, Xylanase and alpha-amylase inhibitor protein isoform III | | Authors: | Singh, A, Kumar, S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-10-08 | | Release date: | 2013-10-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of allergen protein scam1 from Scadoxus multiflorus
To be published
|
|
2Q6N
 
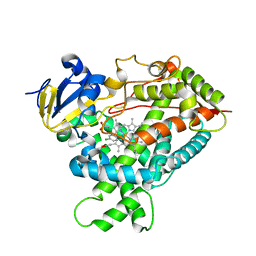 | | Structure of Cytochrome P450 2B4 with Bound 1-(4-cholorophenyl)imidazole | | Descriptor: | 1-(4-CHLOROPHENYL)-1H-IMIDAZOLE, Cytochrome P450 2B4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Zhao, Y, Sun, L, Muralidhara, B.K, Kumar, S, White, M.A, Stout, C.D, Halpert, J.R. | | Deposit date: | 2007-06-05 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural and thermodynamic consequences of 1-(4-chlorophenyl)imidazole binding to cytochrome P450 2B4.
Biochemistry, 46, 2007
|
|
8SR9
 
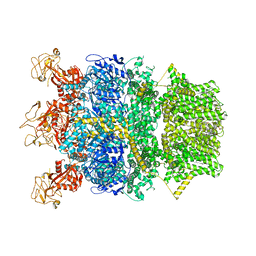 | | Cryo-EM structure of TRPM2 chanzyme in the presence of Magnesium | | Descriptor: | CHOLESTEROL, MAGNESIUM ION, TRPM2 chanzyme | | Authors: | Huang, Y, Kumar, S, Lu, W, Du, J. | | Deposit date: | 2023-05-05 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Coupling enzymatic activity and gating in an ancient TRPM chanzyme and its molecular evolution.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8SR8
 
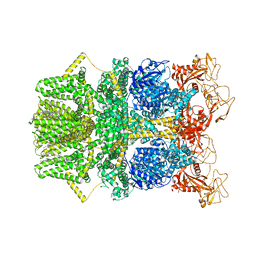 | | Cryo-EM structure of TRPM2 chanzyme in the presence of EDTA (apo state) | | Descriptor: | CHOLESTEROL, TRPM2 chanzyme | | Authors: | Huang, Y, Kumar, S, Lu, W, Du, J. | | Deposit date: | 2023-05-05 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Coupling enzymatic activity and gating in an ancient TRPM chanzyme and its molecular evolution.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8SRA
 
 | | Cryo-EM structure of TRPM2 chanzyme in the presence of Calcium | | Descriptor: | CALCIUM ION, CHOLESTEROL, TRPM2 chanzyme | | Authors: | Huang, Y, Kumar, S, Lu, W, Du, J. | | Deposit date: | 2023-05-05 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Coupling enzymatic activity and gating in an ancient TRPM chanzyme and its molecular evolution.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8SRC
 
 | | Cryo-EM structure of TRPM2 chanzyme in the presence of Calcium and ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, CALCIUM ION, CHOLESTEROL, ... | | Authors: | Huang, Y, Kumar, S, Lu, W, Du, J. | | Deposit date: | 2023-05-05 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Coupling enzymatic activity and gating in an ancient TRPM chanzyme and its molecular evolution.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8SR7
 
 | | Cryo-EM structure of TRPM2 chanzyme in the presence of Magnesium, Adenosine monophosphate, and Ribose-5-phosphate | | Descriptor: | 5-O-phosphono-beta-D-ribofuranose, ADENOSINE MONOPHOSPHATE, CHOLESTEROL, ... | | Authors: | Huang, Y, Kumar, S, Lu, W, Du, J. | | Deposit date: | 2023-05-05 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (1.97 Å) | | Cite: | Coupling enzymatic activity and gating in an ancient TRPM chanzyme and its molecular evolution.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8SRB
 
 | | Cryo-EM structure of TRPM2 chanzyme in the presence of EDTA and ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, CHOLESTEROL, TRPM2 chanzyme | | Authors: | Huang, Y, Kumar, S, Lu, W, Du, J. | | Deposit date: | 2023-05-05 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Coupling enzymatic activity and gating in an ancient TRPM chanzyme and its molecular evolution.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8SRH
 
 | | Cryo-EM structure of TRPM2 chanzyme (E1114A) in the presence of Magnesium and ADP-ribose, open state | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, CHOLESTEROL, MAGNESIUM ION, ... | | Authors: | Huang, Y, Kumar, S, Lu, W, Du, J. | | Deposit date: | 2023-05-05 | | Release date: | 2024-05-15 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Coupling enzymatic activity and gating in an ancient TRPM chanzyme and its molecular evolution.
Nat.Struct.Mol.Biol., 31, 2024
|
|
