7T83
 
 | |
4QKX
 
 | | Structure of beta2 adrenoceptor bound to a covalent agonist and an engineered nanobody | | Descriptor: | 4-[(1R)-1-hydroxy-2-({2-[3-methoxy-4-(2-sulfanylethoxy)phenyl]ethyl}amino)ethyl]benzene-1,2-diol, Beta-2 adrenergic receptor, R9 protein, ... | | Authors: | Weichert, D, Kruse, A.C, Manglik, A, Hiller, C, Zhang, C, Huebner, H, Kobilka, B.K, Gmeiner, P. | | Deposit date: | 2014-06-10 | | Release date: | 2014-07-23 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Covalent agonists for studying G protein-coupled receptor activation.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3UON
 
 | | Structure of the human M2 muscarinic acetylcholine receptor bound to an antagonist | | Descriptor: | (3R)-1-azabicyclo[2.2.2]oct-3-yl hydroxy(diphenyl)acetate, CHLORIDE ION, Human M2 muscarinic acetylcholine, ... | | Authors: | Haga, K, Kruse, A.C, Asada, H, Yurugi-Kobayashi, T, Shiroishi, M, Zhang, C, Weis, W.I, Okada, T, Kobilka, B.K, Haga, T, Kobayashi, T. | | Deposit date: | 2011-11-16 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the human M2 muscarinic acetylcholine receptor bound to an antagonist.
Nature, 482, 2012
|
|
4EJ4
 
 | | Structure of the delta opioid receptor bound to naltrindole | | Descriptor: | (4bS,8R,8aS,14bR)-7-(cyclopropylmethyl)-5,6,7,8,14,14b-hexahydro-4,8-methano[1]benzofuro[2,3-a]pyrido[4,3-b]carbazole-1,8a(9H)-diol, Delta-type opioid receptor, Lysozyme chimera | | Authors: | Granier, S, Manglik, A, Kruse, A.C, Kobilka, T.S, Thian, F.S, Weis, W.I, Kobilka, B.K. | | Deposit date: | 2012-04-06 | | Release date: | 2012-05-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure of the delta opioid receptor bound to naltrindole
Nature, 485, 2012
|
|
4LDO
 
 | | Structure of beta2 adrenoceptor bound to adrenaline and an engineered nanobody | | Descriptor: | (2S)-2,3-dihydroxypropyl (7Z)-tetradec-7-enoate, Camelid Antibody Fragment, L-EPINEPHRINE, ... | | Authors: | Ring, A.M, Manglik, A, Kruse, A.C, Enos, M.D, Weis, W.I, Garcia, K.C, Kobilka, B.K. | | Deposit date: | 2013-06-24 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Adrenaline-activated structure of beta 2-adrenoceptor stabilized by an engineered nanobody.
Nature, 502, 2013
|
|
4LDE
 
 | | Structure of beta2 adrenoceptor bound to BI167107 and an engineered nanobody | | Descriptor: | (2S)-2,3-dihydroxypropyl (7Z)-tetradec-7-enoate, 8-[(1R)-2-{[1,1-dimethyl-2-(2-methylphenyl)ethyl]amino}-1-hydroxyethyl]-5-hydroxy-2H-1,4-benzoxazin-3(4H)-one, Camelid Antibody Fragment, ... | | Authors: | Ring, A.M, Manglik, A, Kruse, A.C, Enos, M.D, Weis, W.I, Garcia, K.C, Kobilka, B.K. | | Deposit date: | 2013-06-24 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Adrenaline-activated structure of beta 2-adrenoceptor stabilized by an engineered nanobody.
Nature, 502, 2013
|
|
4JQI
 
 | | Structure of active beta-arrestin1 bound to a G protein-coupled receptor phosphopeptide | | Descriptor: | 1,2-ETHANEDIOL, Beta-arrestin-1, CHLORIDE ION, ... | | Authors: | Shukla, A.K, Manglik, A, Kruse, A.C, Xiao, K, Reis, R.I, Tseng, W.C, Staus, D.P, Hilger, D, Uysal, S, Huang, L.H, Paduch, M, Shukla, P.T, Koide, A, Koide, S, Weis, W.I, Kossiakoff, A.A, Kobilka, B.K, Lefkowitz, R.J. | | Deposit date: | 2013-03-20 | | Release date: | 2013-04-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of active beta-arrestin-1 bound to a G-protein-coupled receptor phosphopeptide.
Nature, 497, 2013
|
|
4LDL
 
 | | Structure of beta2 adrenoceptor bound to hydroxybenzylisoproterenol and an engineered nanobody | | Descriptor: | (2S)-2,3-dihydroxypropyl (7Z)-tetradec-7-enoate, 4-[(1R)-1-hydroxy-2-{[1-(4-hydroxyphenyl)-2-methylpropan-2-yl]amino}ethyl]benzene-1,2-diol, Camelid Antibody Fragment, ... | | Authors: | Ring, A.M, Manglik, A, Kruse, A.C, Enos, M.D, Weis, W.I, Garcia, K.C, Kobilka, B.K. | | Deposit date: | 2013-06-24 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Adrenaline-activated structure of beta 2-adrenoceptor stabilized by an engineered nanobody.
Nature, 502, 2013
|
|
5HK2
 
 | | Human sigma-1 receptor bound to 4-IBP | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, N-(1-benzylpiperidin-4-yl)-4-iodobenzamide, SULFATE ION, ... | | Authors: | Schmidt, H.R, Zheng, S, Gurpinar, E.G, Koehl, A, Manglik, A, Kruse, A.C. | | Deposit date: | 2016-01-13 | | Release date: | 2016-04-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the human sigma 1 receptor.
Nature, 532, 2016
|
|
5HK1
 
 | | Human sigma-1 receptor bound to PD144418 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3-(4-methylphenyl)-5-(1-propyl-3,6-dihydro-2H-pyridin-5-yl)-1,2-oxazole, SULFATE ION, ... | | Authors: | Schmidt, H.R, Zheng, S, Gurpinar, E, Koehl, A, Manglik, A, Kruse, A.C. | | Deposit date: | 2016-01-13 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5051 Å) | | Cite: | Crystal structure of the human sigma 1 receptor.
Nature, 532, 2016
|
|
3SN6
 
 | | Crystal structure of the beta2 adrenergic receptor-Gs protein complex | | Descriptor: | 8-[(1R)-2-{[1,1-dimethyl-2-(2-methylphenyl)ethyl]amino}-1-hydroxyethyl]-5-hydroxy-2H-1,4-benzoxazin-3(4H)-one, Camelid antibody VHH fragment, Endolysin,Beta-2 adrenergic receptor, ... | | Authors: | Rasmussen, S.G.F, DeVree, B.T, Zou, Y, Kruse, A.C, Chung, K.Y, Kobilka, T.S, Thian, F.S, Chae, P.S, Pardon, E, Calinski, D, Mathiesen, J.M, Shah, S.T.A, Lyons, J.A, Caffrey, M, Gellman, S.H, Steyaert, J, Skiniotis, G, Weis, W.I, Sunahara, R.K, Kobilka, B.K. | | Deposit date: | 2011-06-28 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the beta2 adrenergic receptor-Gs protein complex
Nature, 477, 2011
|
|
6BAR
 
 | | Crystal structure of Thermus thermophilus Rod shape determining protein RodA (Q5SIX3_THET8) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, Rod shape determining protein RodA | | Authors: | Sjodt, M, Brock, K, Dobihal, G, Rohs, P.D.A, Green, A.G, Hopf, T.A, Meeske, A.J, Marks, D.S, Bernhardt, T.G, Rudner, D.Z, Kruse, A.C. | | Deposit date: | 2017-10-15 | | Release date: | 2018-03-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.908 Å) | | Cite: | Structure of the peptidoglycan polymerase RodA resolved by evolutionary coupling analysis.
Nature, 556, 2018
|
|
6BAS
 
 | | Crystal structure of Thermus thermophilus Rod shape determining protein RodA D255A mutant (Q5SIX3_THET8) | | Descriptor: | CHLORIDE ION, Peptidoglycan glycosyltransferase RodA | | Authors: | Sjodt, M, Brock, K, Dobihal, G, Rohs, P.D.A, Green, A.G, Hopf, T.A, Meeske, A.J, Marks, D.S, Bernhardt, T.G, Rudner, D.Z, Kruse, A.C. | | Deposit date: | 2017-10-15 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.194 Å) | | Cite: | Structure of the peptidoglycan polymerase RodA resolved by evolutionary coupling analysis.
Nature, 556, 2018
|
|
6CC4
 
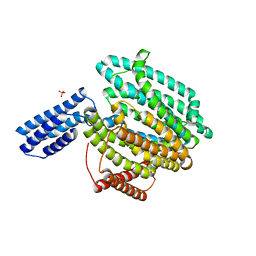 | | Structure of MurJ from Escherichia coli | | Descriptor: | PHOSPHATE ION, soluble cytochrome b562, lipid II flippase MurJ chimera | | Authors: | Zheng, S, Kruse, A.C. | | Deposit date: | 2018-02-05 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure and mutagenic analysis of the lipid II flippase MurJ fromEscherichia coli.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6D35
 
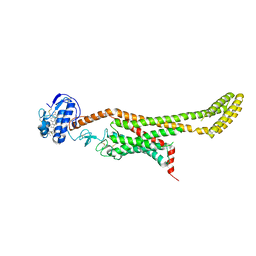 | | Crystal structure of Xenopus Smoothened in complex with cholesterol | | Descriptor: | CHOLESTEROL, Smoothened,Soluble cytochrome b562,Smoothened | | Authors: | Huang, P, Zheng, S, Kim, Y, Kruse, A.C, Salic, A. | | Deposit date: | 2018-04-14 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structural Basis of Smoothened Activation in Hedgehog Signaling.
Cell, 174, 2018
|
|
6D32
 
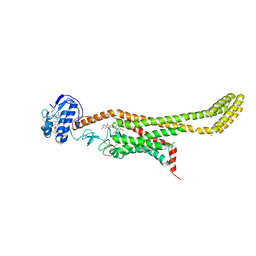 | | Crystal structure of Xenopus Smoothened in complex with cyclopamine | | Descriptor: | Cyclopamine, Smoothened,Soluble cytochrome b562,Smoothened | | Authors: | Huang, P, Zheng, S, Kim, Y, Kruse, A.C, Salic, A. | | Deposit date: | 2018-04-14 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.751 Å) | | Cite: | Structural Basis of Smoothened Activation in Hedgehog Signaling.
Cell, 174, 2018
|
|
6DK1
 
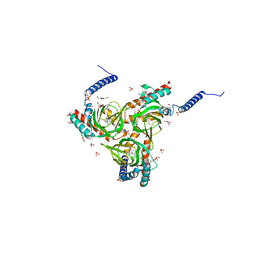 | | Human sigma-1 receptor bound to (+)-pentazocine | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S,6S,11S)-6,11-dimethyl-3-(3-methylbut-2-en-1-yl)-1,2,3,4,5,6-hexahydro-2,6-methano-3-benzazocin-8-ol, GLYCEROL, ... | | Authors: | Schmidt, H.R, Kruse, A.C. | | Deposit date: | 2018-05-28 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Structural basis for sigma1receptor ligand recognition.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DO1
 
 | | Structure of nanobody-stabilized angiotensin II type 1 receptor bound to S1I8 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-like peptide S1I8, ... | | Authors: | Wingler, L.M, McMahon, C, Staus, D.P, Lefkowitz, R.J, Kruse, A.C. | | Deposit date: | 2018-06-08 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | Distinctive Activation Mechanism for Angiotensin Receptor Revealed by a Synthetic Nanobody.
Cell, 176, 2019
|
|
6DK0
 
 | | Human sigma-1 receptor bound to NE-100 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, GLYCEROL, N-{2-[4-methoxy-3-(2-phenylethoxy)phenyl]ethyl}-N-propylpropan-1-amine, ... | | Authors: | Schmidt, H.R, Kruse, A.C. | | Deposit date: | 2018-05-28 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for sigma1receptor ligand recognition.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DJZ
 
 | | Human sigma-1 receptor bound to haloperidol | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-[4-(4-chlorophenyl)-4-hydroxypiperidin-1-yl]-1-(4-fluorophenyl)butan-1-one, GLYCEROL, ... | | Authors: | Schmidt, H.R, Kruse, A.C. | | Deposit date: | 2018-05-28 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.084 Å) | | Cite: | Structural basis for sigma1receptor ligand recognition.
Nat. Struct. Mol. Biol., 25, 2018
|
|
