2PW3
 
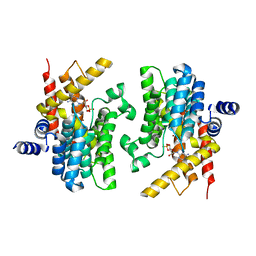 | | Structure of the PDE4D-cAMP complex | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, ZINC ION, cAMP-specific 3',5'-cyclic phosphodiesterase 4D | | Authors: | Wang, H, Robinson, H, Ke, H. | | Deposit date: | 2007-05-10 | | Release date: | 2007-10-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | The molecular basis for different recognition of substrates by phosphodiesterase families 4 and 10.
J.Mol.Biol., 371, 2007
|
|
3ECN
 
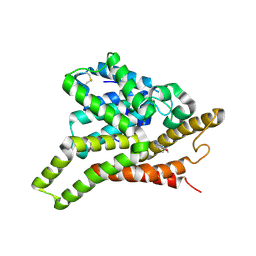 | | Crystal structure of PDE8A catalytic domain in complex with IBMX | | Descriptor: | 3-ISOBUTYL-1-METHYLXANTHINE, High affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8A, MAGNESIUM ION, ... | | Authors: | Wang, H, Yan, Z, Yang, S, Cai, J, Robinson, H, Ke, H. | | Deposit date: | 2008-09-01 | | Release date: | 2008-11-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Kinetic and structural studies of phosphodiesterase-8A and implication on the inhibitor selectivity
Biochemistry, 47, 2008
|
|
3ECM
 
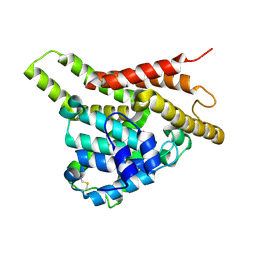 | | Crystal structure of the unliganded PDE8A catalytic domain | | Descriptor: | High affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8A, MAGNESIUM ION, ZINC ION | | Authors: | Wang, H, Yan, Z, Yang, S, Cai, J, Robinson, H, Ke, H. | | Deposit date: | 2008-09-01 | | Release date: | 2008-11-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Kinetic and structural studies of phosphodiesterase-8A and implication on the inhibitor selectivity
Biochemistry, 47, 2008
|
|
6CBU
 
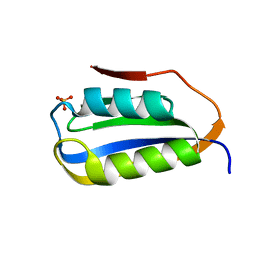 | | Crystal structure of C4S3: A computationally designed immunogen to target Carbohydrate-Occluded Epitopes on the HIV envelope | | Descriptor: | Acylphosphatase-1, SULFATE ION | | Authors: | Zhu, C, Ke, H.M, Swanstrom, R, Dokholyan, N.V. | | Deposit date: | 2018-02-05 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Rationally designed carbohydrate-occluded epitopes elicit HIV-1 Env-specific antibodies.
Nat Commun, 10, 2019
|
|
6CFE
 
 | |
5ED9
 
 | | Crystal structure of CC1 of mouse SUN2 | | Descriptor: | SUN domain-containing protein 2 | | Authors: | Nie, S, Ke, H.M, Gao, F, Ren, J.Q, Wang, M.Z, Huo, L, Gong, W.M, Feng, W. | | Deposit date: | 2015-10-21 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.009 Å) | | Cite: | Coiled-Coil Domains of SUN Proteins as Intrinsic Dynamic Regulators
Structure, 24, 2016
|
|
5ED8
 
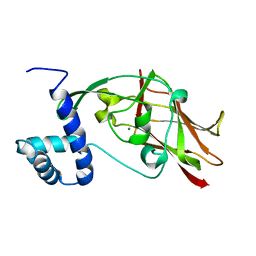 | | Crystal structure of CC2-SUN of mouse SUN2 | | Descriptor: | MAGNESIUM ION, MKIAA0668 protein | | Authors: | Nie, S, Ke, H.M, Gao, F, Ren, J.Q, Wang, M.Z, Huo, L, Gong, W.M, Feng, W. | | Deposit date: | 2015-10-21 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Coiled-Coil Domains of SUN Proteins as Intrinsic Dynamic Regulators
Structure, 24, 2016
|
|
1Y4S
 
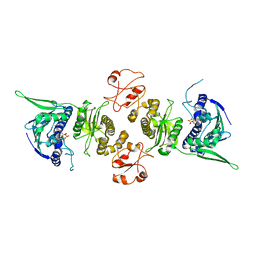 | | Conformation rearrangement of heat shock protein 90 upon ADP binding | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperone protein htpG, MAGNESIUM ION | | Authors: | Huai, Q, Wang, H, Liu, Y, Kim, H, Toft, D, Ke, H. | | Deposit date: | 2004-12-01 | | Release date: | 2005-04-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of the N-terminal and middle domains of E. coli Hsp90 and conformation changes upon ADP binding.
Structure, 13, 2005
|
|
1Y4U
 
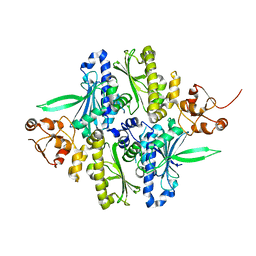 | | Conformation rearrangement of heat shock protein 90 upon ADP binding | | Descriptor: | Chaperone protein htpG | | Authors: | Huai, Q, Wang, H, Liu, Y, Kim, H, Toft, D, Ke, H. | | Deposit date: | 2004-12-01 | | Release date: | 2005-04-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of the N-terminal and middle domains of E. coli Hsp90 and conformation changes upon ADP binding.
Structure, 13, 2005
|
|
1ZKN
 
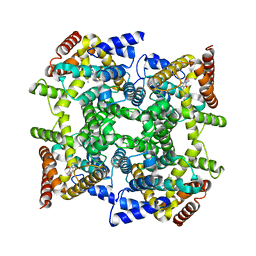 | | Structure of PDE4D2-IBMX | | Descriptor: | 3-ISOBUTYL-1-METHYLXANTHINE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Huai, Q, Liu, Y, Francis, S.H, Corbin, J.D, Ke, H. | | Deposit date: | 2005-05-03 | | Release date: | 2005-05-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Phosphodiesterases 4 and 5 in Complex with Inhibitor 3-Isobutyl-1-Methylxanthine Suggest a Conformation Determinant of Inhibitor Selectivity
J.Biol.Chem., 279, 2004
|
|
1ZKL
 
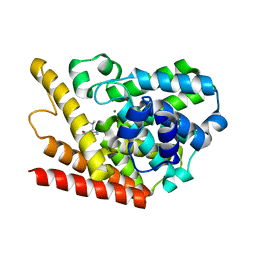 | | Multiple Determinants for Inhibitor Selectivity of Cyclic Nucleotide Phosphodiesterases | | Descriptor: | 3-ISOBUTYL-1-METHYLXANTHINE, High-affinity cAMP-specific 3',5'-cyclic phosphodiesterase 7A, MAGNESIUM ION, ... | | Authors: | Wang, H, Liu, Y, Chen, Y, Robinson, H, Ke, H. | | Deposit date: | 2005-05-03 | | Release date: | 2005-07-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Multiple elements jointly determine inhibitor selectivity of cyclic nucleotide phosphodiesterases 4 and 7
J.Biol.Chem., 280, 2005
|
|
2RBL
 
 | |
2RB8
 
 | |
2R8Q
 
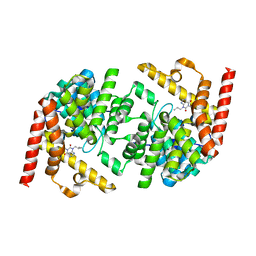 | | Structure of LmjPDEB1 in complex with IBMX | | Descriptor: | 3-ISOBUTYL-1-METHYLXANTHINE, Class I phosphodiesterase PDEB1, MAGNESIUM ION, ... | | Authors: | Wang, H, Yan, Z, Geng, J, Kunz, S, Seebeck, T, Ke, H. | | Deposit date: | 2007-09-11 | | Release date: | 2007-12-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the Leishmania major phosphodiesterase LmjPDEB1 and insight into the design of the parasite-selective inhibitors.
Mol.Microbiol., 66, 2007
|
|
1IAX
 
 | | CRYSTAL STRUCTURE OF ACC SYNTHASE COMPLEXED WITH PLP | | Descriptor: | 1-AMINOCYCLOPROPANE-1-CARBOXYLATE SYNTHASE 2, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Huai, Q, Xia, Y, Chen, Y, Callahan, B, Li, N, Ke, H. | | Deposit date: | 2001-03-24 | | Release date: | 2001-04-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of 1-aminocyclopropane-1-carboxylate (ACC) synthase in complex with aminoethoxyvinylglycine and pyridoxal-5'-phosphate provide new insight into catalytic mechanisms
J.Biol.Chem., 276, 2001
|
|
3QI3
 
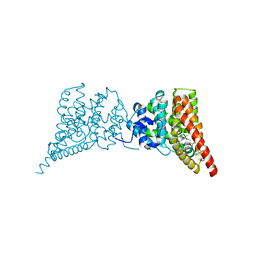 | | Crystal structure of PDE9A(Q453E) in complex with inhibitor BAY73-6691 | | Descriptor: | 1-(2-chlorophenyl)-6-[(2R)-3,3,3-trifluoro-2-methylpropyl]-1,7-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, ... | | Authors: | Hou, J, Xu, J, Liu, M, Zhao, R, Lou, H, Ke, H. | | Deposit date: | 2011-01-26 | | Release date: | 2011-04-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural asymmetry of phosphodiesterase-9, potential protonation of a glutamic Acid, and role of the invariant glutamine.
Plos One, 6, 2011
|
|
3K3E
 
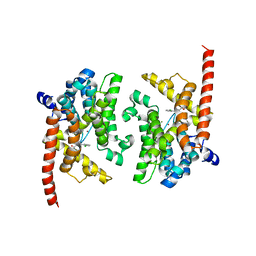 | | Crystal structure of the PDE9A catalytic domain in complex with (R)-BAY73-6691 | | Descriptor: | 1-(2-chlorophenyl)-6-[(2R)-3,3,3-trifluoro-2-methylpropyl]-1,7-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, ... | | Authors: | Wang, H, Luo, X, Ye, M, Hou, J, Robinson, H, Ke, H. | | Deposit date: | 2009-10-02 | | Release date: | 2010-02-16 | | Last modified: | 2013-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Insight into Binding of Phosphodiesterase-9A Selective Inhibitors by Crystal Structures and Mutagenesis
J.Med.Chem., 53, 2010
|
|
3K3H
 
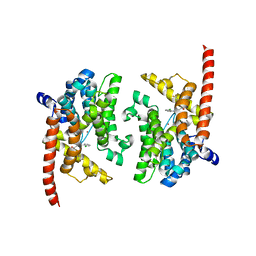 | | Crystal structure of the PDE9A catalytic domain in complex with (S)-BAY73-6691 | | Descriptor: | 1-(2-chlorophenyl)-6-[(2S)-3,3,3-trifluoro-2-methylpropyl]-1,7-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, ... | | Authors: | Wang, H, Luo, X, Ye, M, Hou, J, Robinson, H, Ke, H. | | Deposit date: | 2009-10-02 | | Release date: | 2010-02-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insight into Binding of Phosphodiesterase-9A Selective Inhibitors by Crystal Structures and Mutagenesis
J.Med.Chem., 53, 2010
|
|
1IAW
 
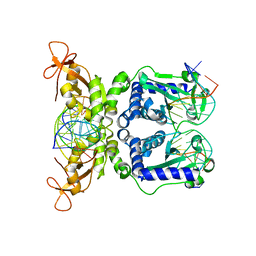 | | CRYSTAL STRUCTURE OF NAEI COMPLEXED WITH 17MER DNA | | Descriptor: | 5'-D(*TP*GP*CP*CP*AP*CP*GP*CP*CP*GP*GP*CP*GP*TP*GP*GP*C)-3', TYPE II RESTRICTION ENZYME NAEI | | Authors: | Huai, Q, Colandene, J.D, Topal, M.D, Ke, H. | | Deposit date: | 2001-03-23 | | Release date: | 2001-08-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of NaeI-DNA complex reveals dual-mode DNA recognition and complete dimer rearrangement.
Nat.Struct.Biol., 8, 2001
|
|
4QGE
 
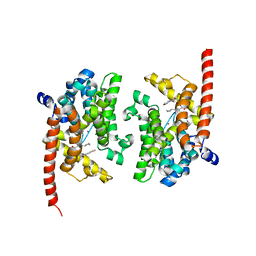 | | phosphodiesterase-9A in complex with inhibitor WYQ-C36D | | Descriptor: | MAGNESIUM ION, N~2~-(1-cyclopentyl-4-oxo-4,7-dihydro-1H-pyrazolo[3,4-d]pyrimidin-6-yl)-N-(4-methoxyphenyl)-D-alaninamide, Phosphodiesterase 9A, ... | | Authors: | Shao, Y.-X, Huang, M, Cui, W, Ke, H. | | Deposit date: | 2014-05-22 | | Release date: | 2014-12-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of a Phosphodiesterase 9A Inhibitor as a Potential Hypoglycemic Agent.
J.Med.Chem., 57, 2014
|
|
3QI4
 
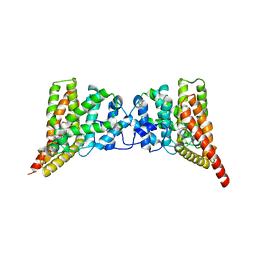 | | Crystal structure of PDE9A(Q453E) in complex with IBMX | | Descriptor: | 3-ISOBUTYL-1-METHYLXANTHINE, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, ... | | Authors: | Hou, J, Xu, J, Liu, M, Zhao, R, Lou, H, Ke, H. | | Deposit date: | 2011-01-26 | | Release date: | 2011-04-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural asymmetry of phosphodiesterase-9, potential protonation of a glutamic Acid, and role of the invariant glutamine.
Plos One, 6, 2011
|
|
1F0J
 
 | | CATALYTIC DOMAIN OF HUMAN PHOSPHODIESTERASE 4B2B | | Descriptor: | ARSENIC, MAGNESIUM ION, PHOSPHODIESTERASE 4B, ... | | Authors: | Xu, R.X, Hassell, A.M, Vanderwall, D, Lambert, M.H, Holmes, W.D, Luther, M.A, Rocque, W.J, Milburn, M.V, Zhao, Y, Ke, H, Nolte, R.T. | | Deposit date: | 2000-05-16 | | Release date: | 2000-07-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Atomic structure of PDE4: insights into phosphodiesterase mechanism and specificity.
Science, 288, 2000
|
|
1COM
 
 | | THE MONOFUNCTIONAL CHORISMATE MUTASE FROM BACILLUS SUBTILIS: STRUCTURE DETERMINATION OF CHORISMATE MUTASE AND ITS COMPLEXES WITH A TRANSITION STATE ANALOG AND PREPHENATE, AND IMPLICATIONS ON THE MECHANISM OF ENZYMATIC REACTION | | Descriptor: | CHORISMATE MUTASE, PREPHENIC ACID | | Authors: | Chook, Y.M, Ke, H, Lipscomb, W.N. | | Deposit date: | 1994-04-08 | | Release date: | 1994-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The monofunctional chorismate mutase from Bacillus subtilis. Structure determination of chorismate mutase and its complexes with a transition state analog and prephenate, and implications for the mechanism of the enzymatic reaction.
J.Mol.Biol., 240, 1994
|
|
4OJV
 
 | | Crystal structure of unliganded yeast PDE1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3',5'-cyclic-nucleotide phosphodiesterase 1, SULFATE ION, ... | | Authors: | Tian, Y, Cui, W, Huang, M, Robinson, H, Wan, Y, Wang, Y, Ke, H. | | Deposit date: | 2014-01-21 | | Release date: | 2014-12-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Dual specificity and novel structural folding of yeast phosphodiesterase-1 for hydrolysis of second messengers cyclic adenosine and guanosine 3',5'-monophosphate.
Biochemistry, 53, 2014
|
|
4OJX
 
 | | crystal structure of yeast phosphodiesterase-1 in complex with GMP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3',5'-cyclic-nucleotide phosphodiesterase 1, GUANOSINE-5'-MONOPHOSPHATE, ... | | Authors: | Tian, Y, Cui, W, Huang, M, Robinson, H, Wan, Y, Wang, Y, Ke, H. | | Deposit date: | 2014-01-21 | | Release date: | 2014-12-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Dual specificity and novel structural folding of yeast phosphodiesterase-1 for hydrolysis of second messengers cyclic adenosine and guanosine 3',5'-monophosphate.
Biochemistry, 53, 2014
|
|
