3KEX
 
 | | Crystal structure of the catalytically inactive kinase domain of the human epidermal growth factor receptor 3 (HER3) | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Receptor tyrosine-protein kinase erbB-3 | | Authors: | Jura, N, Shan, Y, Cao, X, Shaw, D.E, Kuriyan, J. | | Deposit date: | 2009-10-26 | | Release date: | 2009-12-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.797 Å) | | Cite: | Structural analysis of the catalytically inactive kinase domain of the human EGF receptor 3.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3GT8
 
 | | Crystal structure of the inactive EGFR kinase domain in complex with AMP-PNP | | Descriptor: | Epidermal growth factor receptor, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Jura, N, Endres, N.F, Engel, K, Deindl, S, Das, R, Lamers, M.H, Wemmer, D.E, Zhang, X, Kuriyan, J. | | Deposit date: | 2009-03-27 | | Release date: | 2009-07-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.955 Å) | | Cite: | Mechanism for activation of the EGF receptor catalytic domain by the juxtamembrane segment.
Cell(Cambridge,Mass.), 137, 2009
|
|
2KNE
 
 | | Calmodulin wraps around its binding domain in the plasma membrane CA2+ pump anchored by a novel 18-1 motif | | Descriptor: | ATPase, Ca++ transporting, plasma membrane 4, ... | | Authors: | Juranic, N, Atanasova, E, Filoteo, A.G, Macura, S, Prendergast, F.G, Penniston, J.T, Strehler, E.E. | | Deposit date: | 2009-08-21 | | Release date: | 2009-11-24 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Calmodulin wraps around its binding domain in the plasma membrane Ca2+ pump anchored by a novel 18-1 motif.
J.Biol.Chem., 285, 2010
|
|
2KUH
 
 | | Halothane binds to druggable sites in calcium-calmodulin: Solution structure of halothane-CaM C-terminal domain | | Descriptor: | 2-BROMO-2-CHLORO-1,1,1-TRIFLUOROETHANE, CALCIUM ION, Calmodulin | | Authors: | Juranic, N, Macura, S, Simeonov, M.V, Jones, K.A, Penheiter, A.R, Hock, T.J, Streiff, J.H. | | Deposit date: | 2010-02-17 | | Release date: | 2010-03-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Halothane binds to druggable sites in the [Ca2+]4-calmodulin (CaM) complex, but does not inhibit [Ca2+]4-CaM activation of kinase.
J. Serb. Chem. Soc., 78, 2013
|
|
2KUG
 
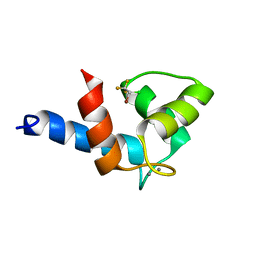 | | Halothane binds to druggable sites in calcium-calmodulin: Solution Structure of halothane-CaM N-terminal domain | | Descriptor: | 2-BROMO-2-CHLORO-1,1,1-TRIFLUOROETHANE, CALCIUM ION, Calmodulin-1 | | Authors: | Juranic, N, Macura, S, Simeonov, M.V, Jones, K.A, Penheiter, A.R, Hock, T.J, Streiff, J.H. | | Deposit date: | 2010-02-17 | | Release date: | 2010-03-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Halothane binds to druggable sites in the [Ca2+]4-calmodulin (CaM) complex, but does not inhibit [Ca2+]4-CaM activation of kinase.
J. Serb. Chem. Soc., 78, 2013
|
|
6OWV
 
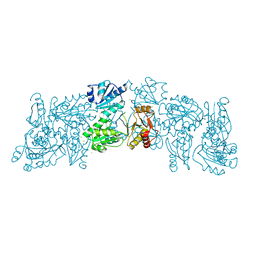 | | Crystal structure of a Human Cardiac Calsequestrin Filament | | Descriptor: | CHLORIDE ION, Calsequestrin-2, SULFATE ION | | Authors: | Titus, E.W, Deiter, F.H, Shi, C, Jura, N, Deo, R.C. | | Deposit date: | 2019-05-12 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The structure of a calsequestrin filament reveals mechanisms of familial arrhythmia.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6OWW
 
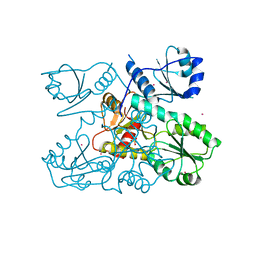 | | Crystal structure of a Human Cardiac Calsequestrin Filament Complexed with Ytterbium | | Descriptor: | Calsequestrin-2, SULFATE ION, YTTERBIUM (III) ION | | Authors: | Titus, E.W, Deiter, F.H, Shi, C, Jura, N, Deo, R.C. | | Deposit date: | 2019-05-12 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.84 Å) | | Cite: | The structure of a calsequestrin filament reveals mechanisms of familial arrhythmia.
Nat.Struct.Mol.Biol., 27, 2020
|
|
5CNO
 
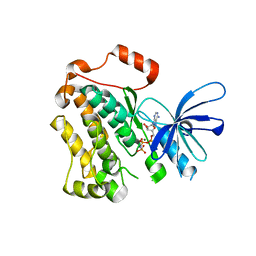 | | Crystal structure of the EGFR kinase domain mutant V924R | | Descriptor: | Epidermal growth factor receptor, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Kovacs, E, Das, R, Mirza, A, Jura, N, Barros, T, Kuriyan, J. | | Deposit date: | 2015-07-17 | | Release date: | 2015-07-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Analysis of the Role of the C-Terminal Tail in the Regulation of the Epidermal Growth Factor Receptor.
Mol.Cell.Biol., 35, 2015
|
|
5CNN
 
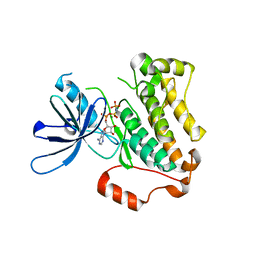 | | Crystal structure of the EGFR kinase domain mutant I682Q | | Descriptor: | Epidermal growth factor receptor, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Kovacs, E, Das, R, Mirza, A, Jura, N, Barros, T, Kuriyan, J. | | Deposit date: | 2015-07-17 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Analysis of the Role of the C-Terminal Tail in the Regulation of the Epidermal Growth Factor Receptor.
Mol.Cell.Biol., 35, 2015
|
|
8DS6
 
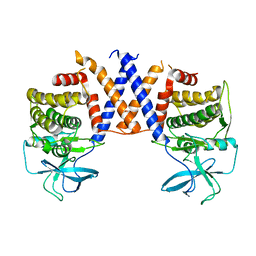 | |
8DP5
 
 | | Structure of the PEAK3/14-3-3 complex | | Descriptor: | 14-3-3 protein beta/alpha, 14-3-3 protein epsilon, Protein PEAK3, ... | | Authors: | Torosyan, H, Paul, M, Jura, N, Verba, K.A. | | Deposit date: | 2022-07-14 | | Release date: | 2023-06-28 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into regulation of the PEAK3 pseudokinase scaffold by 14-3-3.
Nat Commun, 14, 2023
|
|
5WNO
 
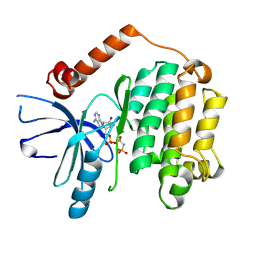 | | Crystal structure of C. elegans LET-23 kinase domain complexed with AMP-PNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Receptor tyrosine-protein kinase let-23 | | Authors: | Liu, L, Thaker, T.M, Jura, N. | | Deposit date: | 2017-08-01 | | Release date: | 2018-01-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.386 Å) | | Cite: | Regulation of Kinase Activity in the Caenorhabditis elegans EGF Receptor, LET-23.
Structure, 26, 2018
|
|
6OP9
 
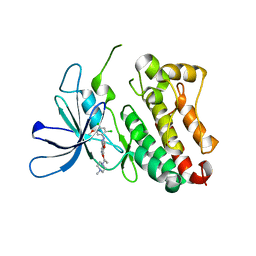 | | HER3 pseudokinase domain bound to bosutinib | | Descriptor: | 4-[(2,4-dichloro-5-methoxyphenyl)amino]-6-methoxy-7-[3-(4-methylpiperazin-1-yl)propoxy]quinoline-3-carbonitrile, Receptor tyrosine-protein kinase erbB-3 | | Authors: | Littlefield, P, Agnew, C, Jura, N. | | Deposit date: | 2019-04-24 | | Release date: | 2019-08-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Targetable HER3 functions driving tumorigenic signaling in HER2-amplified cancers.
Cell Rep, 38, 2022
|
|
6P5S
 
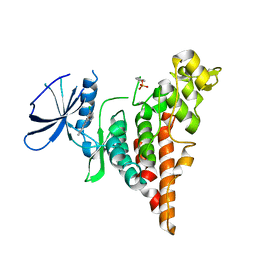 | | HIPK2 kinase domain bound to CX-4945 | | Descriptor: | 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, Homeodomain-interacting protein kinase 2 | | Authors: | Agnew, C, Liu, L, Jura, N. | | Deposit date: | 2019-05-30 | | Release date: | 2019-07-31 | | Last modified: | 2019-09-25 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | The crystal structure of the protein kinase HIPK2 reveals a unique architecture of its CMGC-insert region.
J.Biol.Chem., 294, 2019
|
|
8U4I
 
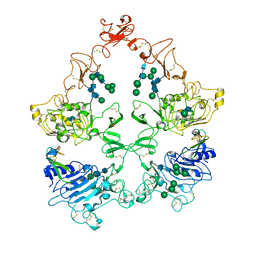 | | Structure of the HER4/NRG1b Homodimer Extracellular Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform 6 of Pro-neuregulin-1, ... | | Authors: | Trenker, R, Diwanji, D, Bingham, T, Verba, K.A, Jura, N. | | Deposit date: | 2023-09-10 | | Release date: | 2024-03-13 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structural dynamics of the active HER4 and HER2/HER4 complexes is finely tuned by different growth factors and glycosylation.
Elife, 12, 2024
|
|
8U4K
 
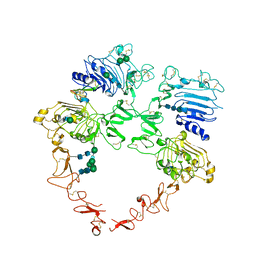 | | Structure of the HER2/HER4/BTC Heterodimer Extracellular Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Betacellulin, ... | | Authors: | Trenker, R, Diwanji, D, Bingham, T, Verba, K.A, Jura, N. | | Deposit date: | 2023-09-10 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.27 Å) | | Cite: | Structural dynamics of the active HER4 and HER2/HER4 complexes is finely tuned by different growth factors and glycosylation.
Elife, 12, 2024
|
|
8U4L
 
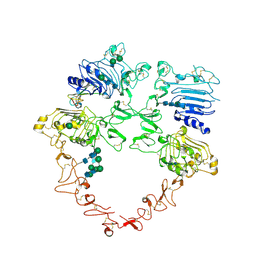 | | Structure of the HER2/HER4/NRG1b Heterodimer Extracellular Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform 6 of Pro-neuregulin-1, ... | | Authors: | Trenker, R, Diwanji, D, Bingham, T, Verba, K.A, Jura, N. | | Deposit date: | 2023-09-10 | | Release date: | 2024-03-13 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Structural dynamics of the active HER4 and HER2/HER4 complexes is finely tuned by different growth factors and glycosylation.
Elife, 12, 2024
|
|
8U4J
 
 | | Structure of the HER4/BTC Homodimer Extracellular Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Betacellulin, ... | | Authors: | Trenker, R, Diwanji, D, Bingham, T, Verba, K.A, Jura, N. | | Deposit date: | 2023-09-10 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural dynamics of the active HER4 and HER2/HER4 complexes is finely tuned by different growth factors and glycosylation.
Elife, 12, 2024
|
|
4RIW
 
 | | Crystal structure of an EGFR/HER3 kinase domain heterodimer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Epidermal growth factor receptor, MAGNESIUM ION, ... | | Authors: | Littlefield, P, Jura, N. | | Deposit date: | 2014-10-07 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural analysis of the EGFR/HER3 heterodimer reveals the molecular basis for activating HER3 mutations.
Sci.Signal., 7, 2014
|
|
4RIY
 
 | | Crystal structure of an EGFR/HER3 kinase domain heterodimer containing the cancer-associated HER3-E909G mutation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Epidermal growth factor receptor, MAGNESIUM ION, ... | | Authors: | Littlefield, P, Liu, L, Jura, N. | | Deposit date: | 2014-10-07 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.981 Å) | | Cite: | Structural analysis of the EGFR/HER3 heterodimer reveals the molecular basis for activating HER3 mutations.
Sci.Signal., 7, 2014
|
|
4RIX
 
 | | Crystal structure of an EGFR/HER3 kinase domain heterodimer containing the cancer-associated HER3-Q790R mutation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Epidermal growth factor receptor, MAGNESIUM ION, ... | | Authors: | Littlefield, P, Liu, L, Jura, N. | | Deposit date: | 2014-10-07 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural analysis of the EGFR/HER3 heterodimer reveals the molecular basis for activating HER3 mutations.
Sci.Signal., 7, 2014
|
|
6CNI
 
 | | Crystal structure of H105A PGAM5 dimer | | Descriptor: | PHOSPHATE ION, SODIUM ION, Serine/threonine-protein phosphatase PGAM5, ... | | Authors: | Ruiz, K, Agnew, C, Jura, N. | | Deposit date: | 2018-03-08 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Functional role of PGAM5 multimeric assemblies and their polymerization into filaments.
Nat Commun, 10, 2019
|
|
6CNL
 
 | | Crystal Structure of H105A PGAM5 Dodecamer | | Descriptor: | MAGNESIUM ION, PGAM5 Multimerization Motif Peptide, Serine/threonine-protein phosphatase PGAM5, ... | | Authors: | Ruiz, K, Agnew, C, Jura, N. | | Deposit date: | 2018-03-08 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Functional role of PGAM5 multimeric assemblies and their polymerization into filaments.
Nat Commun, 10, 2019
|
|
6UNQ
 
 | | Kinase domain of ALK2-K493A with AMPPNP | | Descriptor: | Activin receptor type-1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Agnew, C, Jura, N. | | Deposit date: | 2019-10-13 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for ALK2/BMPR2 receptor complex signaling through kinase domain oligomerization.
Nat Commun, 12, 2021
|
|
6UNR
 
 | | Kinase domain of ALK2-K492A/K493A with AMPPNP | | Descriptor: | Activin receptor type-1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Agnew, C, Jura, N. | | Deposit date: | 2019-10-13 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for ALK2/BMPR2 receptor complex signaling through kinase domain oligomerization.
Nat Commun, 12, 2021
|
|
