7UJD
 
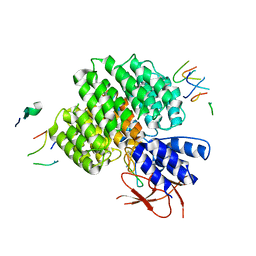 | | PSMD2 Structure bound to MC1 and Fab8/14 | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 2, ACY-PHE-PRO-ASP-VAL-SAR-LEU-HIS-ARG-TYR-TRP-GLY-TRP-ASP-CYS-GLY-NH2, Fab 14 HC CDRs, ... | | Authors: | Johnson, M.C, Bashore, C, Ciferri, C, Dueber, E.C. | | Deposit date: | 2022-03-30 | | Release date: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Targeted degradation via direct 26S proteasome recruitment.
Nat.Chem.Biol., 19, 2023
|
|
7UIH
 
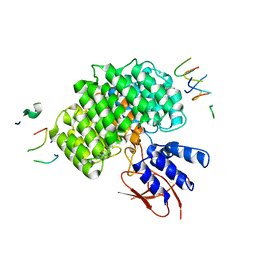 | | PSMD2 Structure | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 2, Fab 14 HC CDRs, Fab 14 LC CDRs, ... | | Authors: | Johnson, M.C, Bashore, C, Ciferri, C, Dueber, E.C. | | Deposit date: | 2022-03-29 | | Release date: | 2023-01-11 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Targeted degradation via direct 26S proteasome recruitment.
Nat.Chem.Biol., 19, 2023
|
|
6U8N
 
 | | Human IMPDH2 treated with ATP, IMP, and NAD+. Fully extended filament segment reconstruction. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase 2, ... | | Authors: | Johnson, M.C, Kollman, J.M. | | Deposit date: | 2019-09-05 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Cryo-EM structures demonstrate human IMPDH2 filament assembly tunes allosteric regulation.
Elife, 9, 2020
|
|
6UDQ
 
 | | Human IMPDH2 treated with ATP, IMP, and 20 mM GTP. Fully compressed filament end reconstruction. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, ... | | Authors: | Johnson, M.C, Kollman, J.M. | | Deposit date: | 2019-09-19 | | Release date: | 2020-03-25 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Cryo-EM structures demonstrate human IMPDH2 filament assembly tunes allosteric regulation.
Elife, 9, 2020
|
|
6U8E
 
 | |
6UDO
 
 | | Human IMPDH2 treated with ATP, IMP, and 20 mM GTP. Fully compressed filament segment reconstruction. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, ... | | Authors: | Johnson, M.C, Kollman, J.M. | | Deposit date: | 2019-09-19 | | Release date: | 2020-03-25 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Cryo-EM structures demonstrate human IMPDH2 filament assembly tunes allosteric regulation.
Elife, 9, 2020
|
|
6UDP
 
 | |
6UA4
 
 | | Human IMPDH2 treated with ATP, IMP, NAD+, and 2 mM GTP. Bent (3/4 compressed, 1/4 extended) segment reconstruction. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, ... | | Authors: | Johnson, M.C, Kollman, J.M. | | Deposit date: | 2019-09-10 | | Release date: | 2020-03-25 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Cryo-EM structures demonstrate human IMPDH2 filament assembly tunes allosteric regulation.
Elife, 9, 2020
|
|
6U8R
 
 | | Human IMPDH2 treated with ATP, IMP, and NAD+. Bent (1/4 compressed, 3/4 extended) segment reconstruction. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase 2, ... | | Authors: | Johnson, M.C, Kollman, J.M. | | Deposit date: | 2019-09-05 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Cryo-EM structures demonstrate human IMPDH2 filament assembly tunes allosteric regulation.
Elife, 9, 2020
|
|
6U9O
 
 | | Human IMPDH2 treated with ATP, IMP, NAD+, and 2 mM GTP. Fully compressed filament segment reconstruction. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, ... | | Authors: | Johnson, M.C, Kollman, J.M. | | Deposit date: | 2019-09-09 | | Release date: | 2020-03-25 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Cryo-EM structures demonstrate human IMPDH2 filament assembly tunes allosteric regulation.
Elife, 9, 2020
|
|
6UA5
 
 | | Human IMPDH2 treated with ATP, IMP, NAD+, and 2 mM GTP. Free interfacial octamer reconstruction. | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase 2, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Johnson, M.C, Kollman, J.M. | | Deposit date: | 2019-09-10 | | Release date: | 2020-03-25 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Cryo-EM structures demonstrate human IMPDH2 filament assembly tunes allosteric regulation.
Elife, 9, 2020
|
|
6U8S
 
 | | Human IMPDH2 treated with ATP, IMP, NAD+, and 2 mM GTP. Filament assembly interface reconstruction. | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase 2, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Johnson, M.C, Kollman, J.M. | | Deposit date: | 2019-09-05 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Cryo-EM structures demonstrate human IMPDH2 filament assembly tunes allosteric regulation.
Elife, 9, 2020
|
|
6UA2
 
 | | Human IMPDH2 treated with ATP, IMP, NAD+, and 2 mM GTP. Bent (2/4 compressed, 2/4 extended) segment reconstruction. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, ... | | Authors: | Johnson, M.C, Kollman, J.M. | | Deposit date: | 2019-09-10 | | Release date: | 2020-03-25 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structures demonstrate human IMPDH2 filament assembly tunes allosteric regulation.
Elife, 9, 2020
|
|
6UAJ
 
 | | Human IMPDH2 treated with ATP, IMP, NAD+, and 2 mM GTP. Free canonical octamer reconstruction. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, ... | | Authors: | Johnson, M.C, Kollman, J.M. | | Deposit date: | 2019-09-10 | | Release date: | 2020-03-25 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Cryo-EM structures demonstrate human IMPDH2 filament assembly tunes allosteric regulation.
Elife, 9, 2020
|
|
6UC2
 
 | |
6OIG
 
 | | Subunit joining exposes nascent pre-40S rRNA for processing and quality control | | Descriptor: | 25S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Rai, J, Parker, M.D, Ghalei, H, Johnson, M.C, Karbstein, K, Stroupe, M.E. | | Deposit date: | 2019-04-09 | | Release date: | 2020-09-30 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | An open interface in the pre-80S ribosome coordinated by ribosome assembly factors Tsr1 and Dim1 enables temporal regulation of Fap7.
Rna, 27, 2021
|
|
8VGI
 
 | |
8VGH
 
 | |
8VGJ
 
 | |
8VGK
 
 | |
7NO8
 
 | | Structure of the mature RSV CA lattice: Group I, pentamer-hexamer interface, class 1"6 | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
7NOM
 
 | | Structure of the mature RSV CA lattice: Group IV, hexamer-hexamer interface, class 4'Beta | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
7NOG
 
 | | Structure of the mature RSV CA lattice: Group III, hexamer-hexamer interface, class 4'5 | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
7NO0
 
 | | Structure of the mature RSV CA lattice: T=1 CA icosahedron | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
7NOB
 
 | | Structure of the mature RSV CA lattice: Group II, hexamer-hexamer interface, class 2'6 | | Descriptor: | Capsid protein p27, alternate cleaved 1 | | Authors: | Obr, M, Ricana, C.L, Nikulin, N, Feathers, J.-P.R, Klanschnig, M, Thader, A, Johnson, M.C, Vogt, V.M, Schur, F.K.M, Dick, R.A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer.
Nat Commun, 12, 2021
|
|
