7SD0
 
 | | Cryo-EM structure of the SHOC2:PP1C:MRAS complex | | Descriptor: | Leucine-rich repeat protein SHOC-2, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Liau, N.P.D, Johnson, M.C, Hymowitz, S.G, Sudhamsu, J. | | Deposit date: | 2021-09-29 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural basis for SHOC2 modulation of RAS signalling.
Nature, 609, 2022
|
|
4NYE
 
 | | Structures of SAICAR Synthetase (PurC) from Streptococcus pneumoniae with ADP, Mg2+, AIR and L-Asp | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Fung, L.W.-M, Johnson, M.E, Abad-Zapatero, C, Wolf, N.M. | | Deposit date: | 2013-12-10 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structures of SAICAR synthetase (PurC) from Streptococcus pneumoniae with ADP, Mg(2+), AIR and Asp.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6MWM
 
 | | Bat coronavirus HKU4 SUD-C | | Descriptor: | Non-structural protein 3 | | Authors: | Staup, A.J, De Silva, I.U, Catt, J.T, Tan, X, Hammond, R.G, Johnson, M.A. | | Deposit date: | 2018-10-29 | | Release date: | 2019-09-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the SARS-Unique Domain C From the Bat Coronavirus HKU4.
Nat Prod Commun, 14, 2019
|
|
3RKZ
 
 | | Discovery of a stable macrocyclic o-aminobenzamide Hsp90 inhibitor capable of significantly decreasing tumor volume in a mouse xenograft model. | | Descriptor: | (5R,6S)-3-(L-alanyl)-5,6,15,15,18-pentamethyl-17-oxo-2,3,4,5,6,7,14,15,16,17-decahydro-1H-12,8-(metheno)[1,5,9]triazacyclotetradecino[1,2-a]indole-9-carboxamide, Heat shock protein HSP 90-alpha | | Authors: | Zapf, C.W, Bloom, J.D, Li, Z, Dushin, R.G, Nittoli, T, Otteng, M, Nikitenko, A, Golas, J.M, Liu, H, Lucas, J, Boschelli, F, Vogan, E, Olland, A, Johnson, M, Levin, J.I. | | Deposit date: | 2011-04-18 | | Release date: | 2011-07-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5693 Å) | | Cite: | Discovery of a stable macrocyclic o-aminobenzamide Hsp90 inhibitor which significantly decreases tumor volume in a mouse xenograft model.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
4PT5
 
 | |
4IQL
 
 | | Crystal Structure of Porphyromonas gingivalis Enoyl-ACP Reductase II (FabK) with cofactors NADPH and FMN | | Descriptor: | Enoyl-(Acyl-carrier-protein) reductase II, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Hevener, K.E, Santarsiero, B.D, Su, P.-C, Boci, T, Truong, K, Johnson, M.E, Mehboob, S. | | Deposit date: | 2013-01-11 | | Release date: | 2014-01-29 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.938 Å) | | Cite: | Structural characterization of Porphyromonas gingivalis enoyl-ACP reductase II (FabK).
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
3MPG
 
 | |
3R5H
 
 | | Crystal Structure of ADP-AIR complex of purK: N5-carboxyaminoimidazole ribonucleotide synthetase | | Descriptor: | 5-AMINOIMIDAZOLE RIBONUCLEOTIDE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Fung, L.W, Tuntland, M.L, Santarsiero, B.D, Johnson, M.E. | | Deposit date: | 2011-03-18 | | Release date: | 2012-03-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Elucidation of the bicarbonate binding site and insights into the carboxylation mechanism of (N(5))-carboxyaminoimidazole ribonucleotide synthase (PurK) from Bacillus anthracis
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3Q2O
 
 | | Crystal Structure of purK: N5-carboxyaminoimidazole ribonucleotide synthetase | | Descriptor: | MAGNESIUM ION, Phosphoribosylaminoimidazole carboxylase, ATPase subunit | | Authors: | Fung, L.W, Tuntland, M.L, Santarsiero, B.D, Johnson, M.E. | | Deposit date: | 2010-12-20 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure of N(5)-carboxyaminoimidazole ribonucleotide synthase (PurK) from Bacillus anthracis.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3QFF
 
 | | Crystal Structure of ADP complex of purK: N5-carboxyaminoimidazole ribonucleotide synthetase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, N5-carboxyaminoimidazole ribonucleotide synthetase | | Authors: | Fung, L.W, Tuntland, M.L, Santarsiero, B.D, Johnson, M.E. | | Deposit date: | 2011-01-21 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Elucidation of the bicarbonate binding site and insights into the carboxylation mechanism of (N(5))-carboxyaminoimidazole ribonucleotide synthase (PurK) from Bacillus anthracis.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3NRC
 
 | | Crystal Structure of the Francisella tularensis enoyl-acyl carrier protein reductase (FabI) in complex with NAD+ and triclosan | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase (NADH), NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRICLOSAN | | Authors: | Mehboob, S, Santarsiero, B.D, Truong, K, Johnson, M.E. | | Deposit date: | 2010-06-30 | | Release date: | 2010-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Structure of the Francisella tularensis enoyl-acyl carrier protein reductase (FabI) in complex with NAD(+) and triclosan.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
4J1N
 
 | | Crystal structures of FabI from F. tularensis in complex with novel inhibitors based on the benzimidazole scaffold | | Descriptor: | 1-(4-methoxy-3-methylbenzyl)-1,5,6,7-tetrahydroindeno[5,6-d]imidazole, Enoyl-[acyl-carrier-protein] reductase [NADH], GLYCEROL, ... | | Authors: | Mehboob, S, Boci, T, Brubaker, L, Santarsiero, B.D, Johnson, M.E. | | Deposit date: | 2013-02-01 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural and biological evaluation of a novel series of benzimidazole inhibitors of Francisella tularensis enoyl-ACP reductase (FabI).
Bioorg.Med.Chem.Lett., 25, 2015
|
|
4NZ9
 
 | | Crystal Structure of FabI from S. aureus in complex with a novel benzimidazole inhibitor | | Descriptor: | 1-(4-methoxy-3-methylbenzyl)-5,6,7,8-tetrahydro-1H-naphtho[2,3-d]imidazole, Enoyl-[acyl-carrier-protein] reductase [NADPH], NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Mehboob, S, Johnson, M.E, Santarsiero, B.D. | | Deposit date: | 2013-12-11 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | To be published
To be Published
|
|
5UTV
 
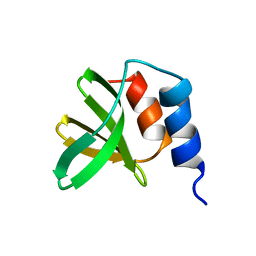 | |
5T1V
 
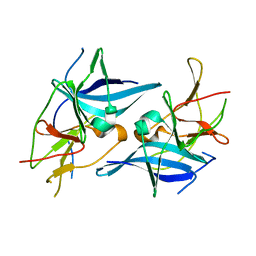 | | Crystal structure of Zika virus NS2B-NS3 protease in apo-form. | | Descriptor: | NS2B-NS3 protease,NS2B-NS3 protease | | Authors: | Nocadello, S, Light, S.H, Minasov, G, Shuvalova, L, Cardona-Correa, A.A, Ojeda, I, Vargas, J, Johnson, M.E, Lee, H, Anderson, W.F, Center for Structural Genomics of Infectious Diseases, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-08-22 | | Release date: | 2016-09-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of Zika virus NS2B-NS3 protease in apo-form.
To Be Published
|
|
5TXG
 
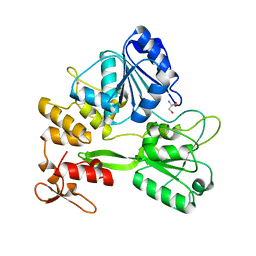 | | Crystal structure of the Zika virus NS3 helicase. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, NS3 helicase, POTASSIUM ION | | Authors: | Nocadello, S, Light, S.H, Minasov, G, Shuvalova, L, Cardona-Correa, A.A, Ojeda, I, Vargas, J, Johnson, M.E, Lee, H, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-16 | | Release date: | 2016-12-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of the Zika virus NS3 helicase.
To be published
|
|
7LBE
 
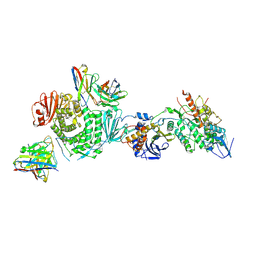 | | CryoEM structure of the HCMV Trimer gHgLgO in complex with neutralizing fabs 13H11 and MSL-109 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein H, ... | | Authors: | Kschonsak, M, Rouge, L, Arthur, C.P, Hoangdung, H, Patel, N, Kim, I, Johnson, M, Kraft, E, Rohou, A.L, Gill, A, Martinez-Martin, N, Payandeh, J, Ciferri, C. | | Deposit date: | 2021-01-07 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of HCMV Trimer reveal the basis for receptor recognition and cell entry.
Cell, 184, 2021
|
|
7LBG
 
 | | CryoEM structure of the HCMV Trimer gHgLgO in complex with human Transforming growth factor beta receptor type 3 and neutralizing fabs 13H11 and MSL-109 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein H, ... | | Authors: | Kschonsak, M, Rouge, L, Arthur, C.P, Hoangdung, H, Patel, N, Kim, I, Johnson, M, Kraft, E, Rohou, A.L, Gill, A, Martinez-Martin, N, Payandeh, J, Ciferri, C. | | Deposit date: | 2021-01-07 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structures of HCMV Trimer reveal the basis for receptor recognition and cell entry.
Cell, 184, 2021
|
|
7LBF
 
 | | CryoEM structure of the HCMV Trimer gHgLgO in complex with human Platelet-derived growth factor receptor alpha and neutralizing fabs 13H11 and MSL-109 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein H, ... | | Authors: | Kschonsak, M, Rouge, L, Arthur, C.P, Hoangdung, H, Patel, N, Kim, I, Johnson, M, Kraft, E, Rohou, A.L, Gill, A, Martinez-Martin, N, Payandeh, J, Ciferri, C. | | Deposit date: | 2021-01-07 | | Release date: | 2021-03-10 | | Last modified: | 2021-03-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures of HCMV Trimer reveal the basis for receptor recognition and cell entry.
Cell, 184, 2021
|
|
8ASJ
 
 | | Four subunit cytochrome b-c1 complex from Rhodobacter sphaeroides in native nanodiscs - focussed refinement in the b-c conformation | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Cytochrome b, Cytochrome b-c1 subunit IV, ... | | Authors: | Swainsbury, D.J.K, Hawkings, F.R, Martin, E.C, Musial, S, Salisbury, J.H, Jackson, P.J, Farmer, D.A, Johnson, M.P, Siebert, C.A, Hitchcock, A, Hunter, C.N. | | Deposit date: | 2022-08-19 | | Release date: | 2023-03-15 | | Last modified: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Cryo-EM structure of the four-subunit Rhodobacter sphaeroides cytochrome bc 1 complex in styrene maleic acid nanodiscs.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8ASI
 
 | | Four subunit cytochrome b-c1 complex from Rhodobacter sphaeroides in native nanodiscs - consensus refinement in the b-b conformation | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Cytochrome b, Cytochrome b-c1 subunit IV, ... | | Authors: | Swainsbury, D.J.K, Hawkings, F.R, Martin, E.C, Musial, S, Salisbury, J.H, Jackson, P.J, Farmer, D.A, Johnson, M.P, Siebert, C.A, Hitchcock, A, Hunter, C.N. | | Deposit date: | 2022-08-19 | | Release date: | 2023-03-15 | | Last modified: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structure of the four-subunit Rhodobacter sphaeroides cytochrome bc 1 complex in styrene maleic acid nanodiscs.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8TLM
 
 | | Structure of a class A GPCR/Fab complex | | Descriptor: | C-C chemokine receptor type 8, Green fluorescent protein fusion, Fab heavy chain, ... | | Authors: | Sun, D, Johnson, M, Masureel, M. | | Deposit date: | 2023-07-27 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of antibody inhibition and chemokine activation of the human CC chemokine receptor 8.
Nat Commun, 14, 2023
|
|
8U1U
 
 | | Structure of a class A GPCR/agonist complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C-C motif chemokine 1,C-C chemokine receptor type 8,EGFP fusion protein, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Sun, D, Johnson, M, Masureel, M. | | Deposit date: | 2023-09-02 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of antibody inhibition and chemokine activation of the human CC chemokine receptor 8.
Nat Commun, 14, 2023
|
|
1M7I
 
 | | CRYSTAL STRUCTURE OF A MONOCLONAL FAB SPECIFIC FOR SHIGELLA FLEXNERI Y LIPOPOLYSACCHARIDE COMPLEXED WITH A PENTASACCHARIDE | | Descriptor: | alpha-L-rhamnopyranose-(1-2)-alpha-L-rhamnopyranose-(1-3)-alpha-L-rhamnopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-methyl 6-deoxy-alpha-L-rhamnopyranoside, heavy chain of the monoclonal antibody Fab SYA/J6, light chain of the monoclonal antibody Fab SYA/J6 | | Authors: | Vyas, N.K, Vyas, M.N, Chervenak, M.C, Johnson, M.A, Pinto, B.M, Bundle, D.R, Quiocho, F.A. | | Deposit date: | 2002-07-19 | | Release date: | 2003-07-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Recognition of Oligosaccharide Epitopes by a Monoclonal Fab Specific for
Shigella flexneri Y Lipopolysaccharide: X-ray Structures and Thermodynamics
Biochemistry, 41, 2002
|
|
4FGR
 
 | | X-Ray Structure of SAICAR Synthetase (PurC) from Streptococcus pneumoniae complexed with ADP and Mg2+ | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Fung, L.W.-M, Johnson, M.E, Abad-Zapatero, C, Wolf, N.W. | | Deposit date: | 2012-06-04 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.597 Å) | | Cite: |
|
|
