6UCR
 
 | | Structure of ClpC1-NTD L92S L96P | | Descriptor: | ACETATE ION, Negative regulator of genetic competence ClpC/mecB | | Authors: | Abad-Zapatero, C, Wolf, N.M. | | Deposit date: | 2019-09-17 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the N-terminal domain of ClpC1 in complex with the antituberculosis natural product ecumicin reveals unique binding interactions.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
7JS3
 
 | | Structure of the Class II Fructose-1,6-Bisphophatase from Francisella tularensis | | Descriptor: | Fructose-1,6-bisphosphatase, GLYCEROL, MAGNESIUM ION | | Authors: | Abad-Zapatero, C, Wolf, N.M, Movahedzadeh, F, Gutka, H, Selezneva, A. | | Deposit date: | 2020-08-13 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and biochemical characterization of the class II fructose-1,6-bisphosphatase from Francisella tularensis.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
4NYE
 
 | | Structures of SAICAR Synthetase (PurC) from Streptococcus pneumoniae with ADP, Mg2+, AIR and L-Asp | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Fung, L.W.-M, Johnson, M.E, Abad-Zapatero, C, Wolf, N.M. | | Deposit date: | 2013-12-10 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structures of SAICAR synthetase (PurC) from Streptococcus pneumoniae with ADP, Mg(2+), AIR and Asp.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6PBA
 
 | | Structure of ClpC1-NTD | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit ClpC1 | | Authors: | Abad-Zapatero, C, Wolf, N.M. | | Deposit date: | 2019-06-13 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structure of the N-terminal domain of ClpC1 in complex with the antituberculosis natural product ecumicin reveals unique binding interactions.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6PBQ
 
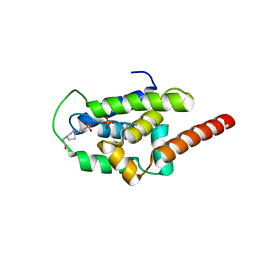 | | Structure of ClpC1-NTD | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ATP-dependent Clp protease ATP-binding subunit ClpC1, PHOSPHATE ION | | Authors: | Abad-Zapatero, C, Wolf, N.M. | | Deposit date: | 2019-06-14 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the N-terminal domain of ClpC1 in complex with the antituberculosis natural product ecumicin reveals unique binding interactions.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6PBS
 
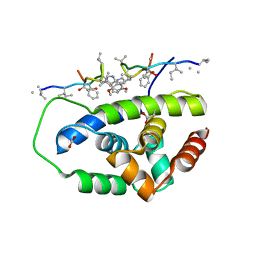 | | Structure of ClpC1-NTD in complex with Ecumicin | | Descriptor: | ACETATE ION, ATP-dependent Clp protease ATP-binding subunit ClpC1, ecumicin | | Authors: | Abad-Zapatero, C, Wolf, N.M. | | Deposit date: | 2019-06-14 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the N-terminal domain of ClpC1 in complex with the antituberculosis natural product ecumicin reveals unique binding interactions.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
