2AFQ
 
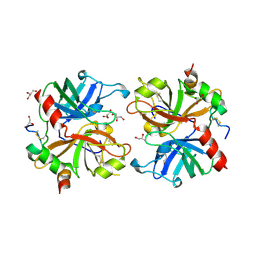 | |
7S53
 
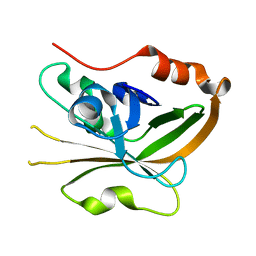 | | Structure of Sortase A from Streptococcus pyogenes with the b7-b8 loop sequence from Listeria monocytogenes Sortase A | | Descriptor: | Class A sortase, sortase A chimera | | Authors: | Johnson, D.A, Svendsen, J.E, Antos, J.M, Amacher, J.F. | | Deposit date: | 2021-09-09 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and biochemical analyses of selectivity determinants in chimeric Streptococcus Class A sortase enzymes.
Protein Sci., 31, 2022
|
|
7S51
 
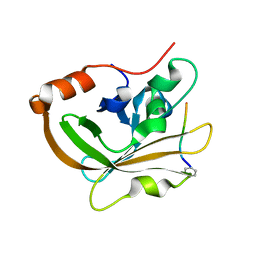 | | Structure of C208A Sortase A from Streptococcus pyogenes bound to LPATA peptide | | Descriptor: | LEU-PRO-ALA-THR-ALA, Sortase | | Authors: | Johnson, D.A, Svendsen, J.E, Antos, J.M, Amacher, J.F. | | Deposit date: | 2021-09-09 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structures of Streptococcus pyogenes class A sortase in complex with substrate and product mimics provide key details of target recognition.
J.Biol.Chem., 298, 2022
|
|
7S4O
 
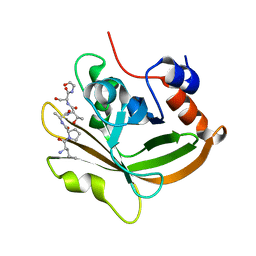 | | Structure of C208A Sortase A from Streptococcus pyogenes bound to LPATS peptide | | Descriptor: | LEU-PRO-ALA-THR-SER-GLY, Sortase | | Authors: | Johnson, D.A, Svendsen, J.E, Antos, J.M, Amacher, J.F. | | Deposit date: | 2021-09-09 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.396 Å) | | Cite: | Structures of Streptococcus pyogenes class A sortase in complex with substrate and product mimics provide key details of target recognition.
J.Biol.Chem., 298, 2022
|
|
1OYH
 
 | | Crystal Structure of P13 Alanine Variant of Antithrombin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antithrombin-III, ... | | Authors: | Johnson, D.J.D, Huntington, J.A. | | Deposit date: | 2003-04-04 | | Release date: | 2004-04-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | The influence of hinge region residue Glu-381 on antithrombin allostery and metastability
J.Biol.Chem., 279, 2004
|
|
1AG5
 
 | | THE SOLUTION STRUCTURE OF AN AFLATOXIN B1 EPOXIDE ADDUCT AT THE N7 POSITION OF GUANINE OPPOSITE AN ADENINE IN THE COMPLEMENTARY STRAND OF AN OLIGODEOXYNUCLEOTIDE DUPLEX, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | 8,9-DIHYDRO-9-HYDROXY-AFLATOXIN B1, DNA (5'-D(*CP*CP*AP*TP*CP*GP*AP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*TP*CP*AP*GP*AP*TP*GP*G)-3') | | Authors: | Johnson, D.S, Stone, M.P. | | Deposit date: | 1997-04-01 | | Release date: | 1997-09-17 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Refined solution structure of 8,9-dihydro-8-(N7-guanyl)-9-hydroxyaflatoxin B1 opposite CpA in the complementary strand of an oligodeoxynucleotide duplex as determined by 1H NMR.
Biochemistry, 34, 1995
|
|
1T1F
 
 | | Crystal Structure of Native Antithrombin in its Monomeric Form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antithrombin-III, GLYCEROL, ... | | Authors: | Johnson, D.J.D, Huntington, J.A. | | Deposit date: | 2004-04-16 | | Release date: | 2005-10-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of monomeric native antithrombin reveals a novel reactive center loop conformation
J.Biol.Chem., 281, 2006
|
|
2GD4
 
 | | Crystal Structure of the Antithrombin-S195A Factor Xa-Pentasaccharide Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-3,6-di-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-methyl 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranoside, ... | | Authors: | Johnson, D.J, Li, W, Adams, T.E, Huntington, J.A. | | Deposit date: | 2006-03-15 | | Release date: | 2006-05-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Antithrombin-S195A factor Xa-heparin structure reveals the allosteric mechanism of antithrombin activation.
Embo J., 25, 2006
|
|
2BEH
 
 | | Crystal structure of antithrombin variant S137A/V317C/T401C with plasma latent antithrombin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antithrombin-III, ... | | Authors: | Johnson, D.J, Luis, S.A, Huntington, J.A. | | Deposit date: | 2005-10-24 | | Release date: | 2005-11-01 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of monomeric native antithrombin reveals a novel reactive center loop conformation.
J.Biol.Chem., 281, 2006
|
|
2B5T
 
 | | 2.1 Angstrom structure of a nonproductive complex between antithrombin, synthetic heparin mimetic SR123781 and two S195A thrombin molecules | | Descriptor: | 2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose-(1-4)-2,3,6-tri-O-sulfo-alpha-D-glucopyranose-(1-4)-2,3,6-tri-O-sulfonato-beta-D-glucopyranose-(1-4)-2,3-di-O-methyl-6-O-sulfonato-alpha-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-beta-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-alpha-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-beta-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-alpha-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-beta-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-alpha-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-beta-D-glucopyranose-(1-4)-2,3-di-O-methyl-6-O-sulfonato-alpha-D-glucopyranose-(1-4)-2,3-di-O-methyl-beta-D-glucopyranuronic acid-(1-4)-2,3,6-tri-O-sulfo-alpha-D-glucopyranose-(1-4)-2,3-di-O-methyl-alpha-L-idopyranuronic acid-(1-4)-methyl 3-O-methyl-2,6-di-O-sulfo-alpha-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Johnson, D.J, Li, W, Luis, S.A, Carrell, R.W, Huntington, J.A. | | Deposit date: | 2005-09-29 | | Release date: | 2006-09-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of monomeric native antithrombin reveals a novel reactive center loop conformation.
J.Biol.Chem., 281, 2006
|
|
3KCG
 
 | | Crystal structure of the antithrombin-factor IXa-pentasaccharide complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3,4-di-O-methyl-2,6-di-O-sulfo-alpha-D-glucopyranose-(1-4)-2,3-di-O-methyl-beta-D-glucopyranuronic acid-(1-4)-2,3,6-tri-O-sulfo-alpha-D-glucopyranose-(1-4)-3-O-methyl-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-methyl 2,3,6-tri-O-sulfo-alpha-D-glucopyranoside, Antithrombin-III, ... | | Authors: | Huntington, J.A, Johnson, D.J.D. | | Deposit date: | 2009-10-21 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular basis of factor IXa recognition by heparin-activated antithrombin revealed by a 1.7-A structure of the ternary complex.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4BXW
 
 | | Crystal Structure of the Prothrombinase Complex from the Venom of Pseudonaja Textilis | | Descriptor: | COAGULATION FACTOR V, FACTOR XA, GLYCEROL, ... | | Authors: | Lechtenberg, B.C, Murray-Rust, T.A, Johnson, D.J.D, Adams, T.E, Krishnaswamy, S, Camire, R.M, Huntington, J.A. | | Deposit date: | 2013-07-16 | | Release date: | 2013-07-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystal Structure of the Prothrombinase Complex from the Venom of Pseudonaja Textilis.
Blood, 122, 2013
|
|
4EB1
 
 | | Hyperstable in-frame insertion variant of antithrombin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antithrombin-III | | Authors: | Martinez-Martinez, I, Johnson, D.J.D, Yamasaki, M, Corral, J, Huntington, J.A. | | Deposit date: | 2012-03-23 | | Release date: | 2012-07-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Type II antithrombin deficiency caused by a large in-frame insertion: structural, functional and pathological relevance.
J.Thromb.Haemost., 10, 2012
|
|
4BXS
 
 | | Crystal Structure of the Prothrombinase Complex from the Venom of Pseudonaja Textilis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Lechtenberg, B.C, Murray-Rust, T.A, Johnson, D.J.D, Adams, T.E, Krishnaswamy, S, Camire, R.M, Huntington, J.A. | | Deposit date: | 2013-07-15 | | Release date: | 2013-07-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Crystal Structure of the Prothrombinase Complex from the Venom of Pseudonaja Textilis.
Blood, 122, 2013
|
|
5MTF
 
 | | A modular route to novel potent and selective inhibitors of rhomboid intramembrane proteases | | Descriptor: | CHLORIDE ION, Rhomboid protease GlpG, inhibitor, ... | | Authors: | Ticha, A, Stanchev, S, Vinothkumar, K.R, Mikles, D.C, Pachl, P, Svehlova, K, Nguyen, M.T.N, Verhelst, S.H.L, Johnson, D, Bachovchin, D, Lepsik, M, Majer, P, Strisovsky, K. | | Deposit date: | 2017-01-09 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | General and Modular Strategy for Designing Potent, Selective, and Pharmacologically Compliant Inhibitors of Rhomboid Proteases.
Cell Chem Biol, 24, 2017
|
|
7RAM
 
 | | Cryo-EM Structure of the HCMV gHgLgO Trimer Derived from AD169 and TR strains in complex with PDGFRalpha | | Descriptor: | Envelope glycoprotein H, Envelope glycoprotein L, Envelope glycoprotein O, ... | | Authors: | Liu, J, Vanarsdall, A.L, Chen, D, Johnson, D.C, Jardetzky, T.S. | | Deposit date: | 2021-07-02 | | Release date: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Cryo-Electron Microscopy Structure and Interactions of the Human Cytomegalovirus gHgLgO Trimer with Platelet-Derived Growth Factor Receptor Alpha.
Mbio, 12, 2021
|
|
7S57
 
 | | Structure of Sortase A from Streptococcus pyogenes with the b7-b8 loop sequence of Enterococcus faecalis Sortase A | | Descriptor: | Class A sortase, sortase A chimera | | Authors: | Svendsen, J.E, Johnson, D.A, Gao, M, Antos, J.M, Amacher, J.F. | | Deposit date: | 2021-09-09 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and biochemical analyses of selectivity determinants in chimeric Streptococcus Class A sortase enzymes.
Protein Sci., 31, 2022
|
|
5NBV
 
 | |
5NBU
 
 | |
1CVW
 
 | | Crystal structure of active site-inhibited human coagulation factor VIIA (DES-GLA) | | Descriptor: | CALCIUM ION, COAGULATION FACTOR VIIA (HEAVY CHAIN) (DES-GLA), COAGULATION FACTOR VIIA (LIGHT CHAIN) (DES-GLA), ... | | Authors: | Kemball-Cook, G, Johnson, D.J.D, Tuddenham, E.G.D, Harlos, K. | | Deposit date: | 1999-08-24 | | Release date: | 1999-08-31 | | Last modified: | 2013-02-27 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of active site-inhibited human coagulation factor VIIa (des-Gla).
J.Struct.Biol., 127, 1999
|
|
3CDG
 
 | | Human CD94/NKG2A in complex with HLA-E | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, alpha chain E, ... | | Authors: | Petrie, E.J, Clements, C.S, Lin, J, Sullivan, L.C, Johnson, D, Huyton, T, Heroux, A, Hoare, H.L, Beddoe, T, Reid, H.H, Wilce, M.C.J, Brooks, A.G, Rossjohn, J. | | Deposit date: | 2008-02-26 | | Release date: | 2008-04-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | CD94-NKG2A recognition of human leukocyte antigen (HLA)-E bound to an HLA class I leader sequence
J.Exp.Med., 205, 2008
|
|
1LPL
 
 | | Structural Genomics of Caenorhabditis elegans: CAP-Gly domain of F53F4.3 | | Descriptor: | Hypothetical 25.4 kDa protein F53F4.3 in chromosome V | | Authors: | Li, S, Finley, J, Liu, Z.-J, Qiu, S.H, Luan, C.H, Carson, M, Tsao, J, Johnson, D, Lin, G, Zhao, J, Thomas, W, Nagy, L.A, Sha, B, DeLucas, L.J, Wang, B.-C, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2002-05-08 | | Release date: | 2002-05-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal Structure of the
Cytoskeleton-associated Protein
Glycine-rich (CAP-Gly) Domain
J.Biol.Chem., 277, 2002
|
|
1TOV
 
 | | Structural genomics of Caenorhabditis elegans: CAP-GLY domain of F53F4.3 | | Descriptor: | Hypothetical protein F53F4.3 in chromosome V, SULFATE ION | | Authors: | Li, S, Finley, J, Liu, Z.J, Qiu, S.H, Luan, C.H, Carson, M, Tsao, J, Johnson, D, Lin, G, Zhao, J, Thomas, W, Nagy, L.A, Sha, B, Delucas, L.J, Richardson, D, Richardson, J, Wang, B.C, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-06-15 | | Release date: | 2004-07-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal Structure of the Cytoskeleton-Associated Protein Glycine-Rich (CAP-Gly) Domain
J.Biol.Chem., 277, 2002
|
|
2VYA
 
 | | Crystal Structure of fatty acid amide hydrolase conjugated with the drug-like inhibitor PF-750 | | Descriptor: | 4-(quinolin-3-ylmethyl)piperidine-1-carboxylic acid, CHLORIDE ION, FATTY-ACID AMIDE HYDROLASE 1, ... | | Authors: | Mileni, M, Johnson, D.S, Wang, Z, Everdeen, D.S, Liimatta, M, Pabst, B, Bhattacharya, K, Nugent, R.A, Kamtekar, S, Cravatt, B.F, Ahn, K, Stevens, R.C. | | Deposit date: | 2008-07-22 | | Release date: | 2008-09-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure-Guided Inhibitor Design for Human Faah by Interspecies Active Site Conversion.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
1NQ9
 
 | | Crystal Structure of Antithrombin in the Pentasaccharide-Bound Intermediate State | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3,4-di-O-methyl-2,6-di-O-sulfo-alpha-D-glucopyranose-(1-4)-2,3-di-O-methyl-beta-D-glucopyranuronic acid-(1-4)-2,3,6-tri-O-sulfo-alpha-D-glucopyranose-(1-4)-3-O-methyl-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-methyl 2,3,6-tri-O-sulfo-alpha-D-glucopyranoside, ... | | Authors: | Huntington, J.A, Johnson, D.J.D. | | Deposit date: | 2003-01-21 | | Release date: | 2003-09-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Antithrombin in a Heparin-Bound Intermediate State
Biochemistry, 42, 2003
|
|
