7EER
 
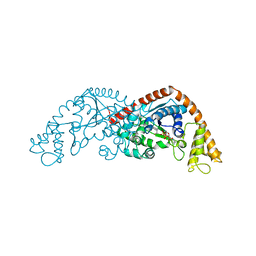 | | Crystal structure of Tryptophanyl-tRNA synthetase from Bacillus stearothermophilus in complex with 05E6 and ATP | | Descriptor: | 2-(1H-indol-3-yl)ethanol, ADENOSINE-5'-TRIPHOSPHATE, Tryptophan--tRNA ligase | | Authors: | Lv, G, Fan, S, Feng, X, Zhang, Q, Wu, G, Jin, Y, Yang, Z. | | Deposit date: | 2021-03-19 | | Release date: | 2022-05-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Tryptophanyl-tRNA synthetase from Bacillus stearothermophilus in complex with O5E6 and ATP
To Be Published
|
|
2A65
 
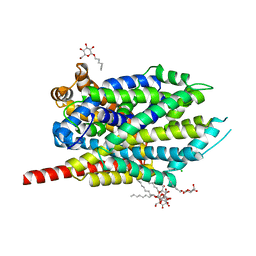 | | Crystal structure of LEUTAA, a bacterial homolog of Na+/Cl--dependent neurotransmitter transporters | | Descriptor: | CHLORIDE ION, LEUCINE, Na(+):neurotransmitter symporter (Snf family), ... | | Authors: | Yamashita, A, Singh, S.K, Kawate, T, Jin, Y, Gouaux, E. | | Deposit date: | 2005-07-01 | | Release date: | 2005-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of a bacterial homologue of Na(+)/Cl(-)-dependent neurotransmitter transporters.
Nature, 437, 2005
|
|
6IXM
 
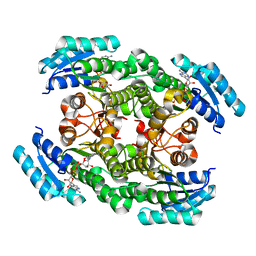 | | Crystal structure of the ketone reductase ChKRED20 from the genome of Chryseobacterium sp. CA49 complexed with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Short-chain dehydrogenase reductase | | Authors: | Zhao, F.J, Jin, Y, Liu, Z.C, Wang, G.G, Wu, Z.L. | | Deposit date: | 2018-12-11 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Structure-guided engineering of ChKRED20 from Chryseobacterium sp. CA49 for asymmetric reduction of aryl ketoesters.
Enzyme Microb. Technol., 125, 2019
|
|
6HG6
 
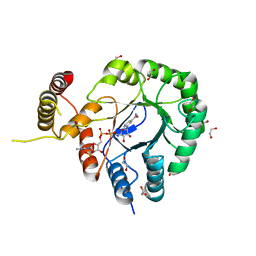 | | Clostridium beijerinckii aldo-keto reductase Cbei_3974 with NADPH | | Descriptor: | 1,2-ETHANEDIOL, L-glyceraldehyde 3-phosphate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Scott, A.F, Cresser-Brown, J, Rizkallah, P.J, Jin, Y, Allemann, R.K. | | Deposit date: | 2018-08-22 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure and Biophysical Analysis of Furfural-Detoxifying Aldehyde Reductase from Clostridium beijerinckii.
Appl.Environ.Microbiol., 85, 2019
|
|
7DVA
 
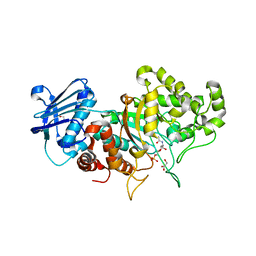 | | Structure of wild type Bt4394, a GH20 family sulfoglycosidase, in complex with 6S-GlcNAc | | Descriptor: | 2-acetamido-2-deoxy-6-O-sulfo-beta-D-glucopyranose, Beta-N-acetylhexosaminidase, GLYCEROL | | Authors: | Zhang, Z, He, Y, Jin, Y. | | Deposit date: | 2021-01-13 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Mechanistic and Structural Insights into the Specificity and Biological Functions of Bacterial Sulfoglycosidases
Acs Catalysis, 13, 2023
|
|
7DUP
 
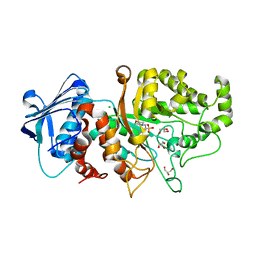 | | Apo structure of wild type Bt4394, a GH20 family sulfoglycosidase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-N-acetylhexosaminidase, CHLORIDE ION, ... | | Authors: | Zhang, Z, He, Y, Jin, Y. | | Deposit date: | 2021-01-10 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Mechanistic and Structural Insights into the Specificity and Biological Functions of Bacterial Sulfoglycosidases
Acs Catalysis, 13, 2023
|
|
7DVB
 
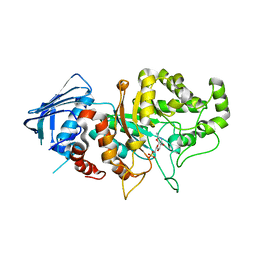 | | D335N variant of Bt4394 in complex with 6SO3-NAG-oxazoline intermediate | | Descriptor: | 2-acetamido-2-deoxy-6-O-sulfo-beta-D-glucopyranose, Beta-N-acetylhexosaminidase, [(3~{a}~{R},5~{R},6~{S},7~{R},7~{a}~{R})-2-methyl-6,7-bis(oxidanyl)-5,6,7,7~{a}-tetrahydro-3~{a}~{H}-pyrano[3,2-d][1,3]oxazol-1-ium-5-yl]methyl sulfate | | Authors: | Zhang, Z, He, Y, Jin, Y. | | Deposit date: | 2021-01-13 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Mechanistic and Structural Insights into the Specificity and Biological Functions of Bacterial Sulfoglycosidases
Acs Catalysis, 13, 2023
|
|
7VNE
 
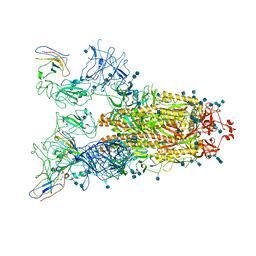 | | Structure of the SARS-CoV-2 spike glycoprotein in complex with a human single domain antibody n3113.1 (UUU-state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yang, Z, Wang, Y, Kong, Y, Jin, Y, Wu, Y, Ying, T. | | Deposit date: | 2021-10-10 | | Release date: | 2021-11-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | A non-ACE2 competing human single-domain antibody confers broad neutralization against SARS-CoV-2 and circulating variants.
Signal Transduct Target Ther, 6, 2021
|
|
1O5T
 
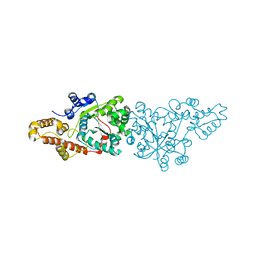 | | Crystal structure of the aminoacylation catalytic fragment of human tryptophanyl-tRNA synthetase | | Descriptor: | Tryptophanyl-tRNA synthetase | | Authors: | Yu, Y, Liu, Y, Shen, N, Xu, X, Jia, J, Jin, Y, Arnold, E, Ding, J. | | Deposit date: | 2003-10-05 | | Release date: | 2004-07-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Human Tryptophanyl-tRNA Synthetase Catalytic Fragment
J.BIOL.CHEM., 279, 2004
|
|
7VNC
 
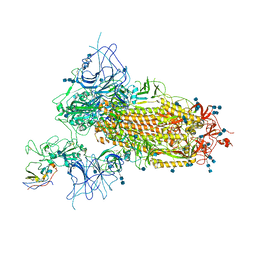 | | Structure of the SARS-CoV-2 spike glycoprotein in complex with a human single domain antibody n3113 (UDD-state, state 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yang, Z, Wang, Y, Kong, Y, Jin, Y, Wu, Y, Ying, T. | | Deposit date: | 2021-10-10 | | Release date: | 2021-11-24 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A non-ACE2 competing human single-domain antibody confers broad neutralization against SARS-CoV-2 and circulating variants.
Signal Transduct Target Ther, 6, 2021
|
|
7VND
 
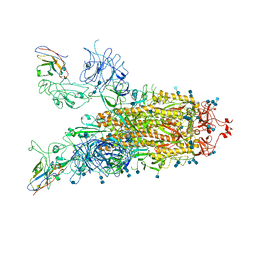 | | Structure of the SARS-CoV-2 spike glycoprotein in complex with a human single domain antibody n3113 (UUD-state, state 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Yang, Z, Wang, Y, Kong, Y, Jin, Y, Wu, Y, Ying, T. | | Deposit date: | 2021-10-10 | | Release date: | 2021-11-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A non-ACE2 competing human single-domain antibody confers broad neutralization against SARS-CoV-2 and circulating variants.
Signal Transduct Target Ther, 6, 2021
|
|
7VNB
 
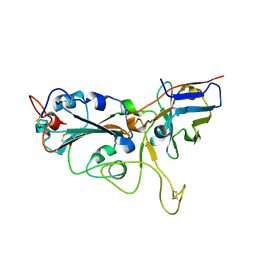 | | Crystal structure of the SARS-CoV-2 RBD in complex with a human single domain antibody n3113 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, n3113 | | Authors: | Yang, Z, Wang, Y, Kong, Y, Jin, Y, Wu, Y, Ying, T. | | Deposit date: | 2021-10-10 | | Release date: | 2021-11-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | A non-ACE2 competing human single-domain antibody confers broad neutralization against SARS-CoV-2 and circulating variants.
Signal Transduct Target Ther, 6, 2021
|
|
7CYI
 
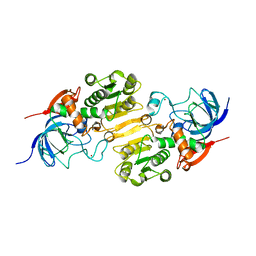 | | Crystal structure of Alcohol dehydrogenase 1 from Artemisia annua | | Descriptor: | Alcohol dehydrogenase 1, ZINC ION | | Authors: | Feng, X, Fan, S, Lv, G, Li, M, Wu, G, Jin, Y, Yang, Z. | | Deposit date: | 2020-09-03 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structure of Alcohol dehydrogenase 1 from Artemisia annua
To Be Published
|
|
7C8U
 
 | | The crystal structure of COVID-19 main protease in complex with GC376 | | Descriptor: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Luan, X, Shang, W, Wang, Y, Yin, W, Jiang, Y, Feng, S, Wang, Y, Liu, M, Zhou, R, Zhang, Z, Wang, F, Cheng, W, Gao, M, Wang, H, Wu, W, Tian, R, Tian, Z, Jin, Y, Jiang, H.W, Zhang, L, Xu, H.E, Zhang, S. | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The crystal structure of COVID-19 main protease in complex with GC376
To Be Published
|
|
6A5L
 
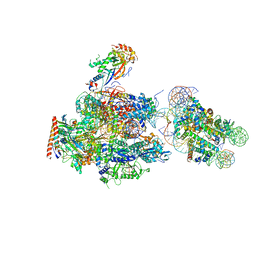 | | RNA polymerase II elongation complex stalled at SHL(-1) of the nucleosome, with foreign DNA | | Descriptor: | DNA (198-MER), DNA (42-MER), DNA-directed RNA polymerase subunit, ... | | Authors: | Kujirai, T, Ehara, H, Fujino, Y, Shirouzu, M, Sekine, S, Kurumizaka, H. | | Deposit date: | 2018-06-24 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Structural basis of the nucleosome transition during RNA polymerase II passage.
Science, 362, 2018
|
|
6A5R
 
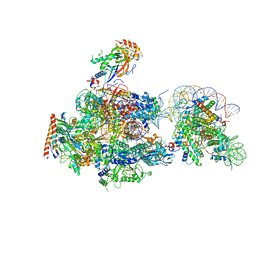 | | RNA polymerase II elongation complex stalled at SHL(-2) of the nucleosome | | Descriptor: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Kujirai, T, Ehara, H, Fujino, Y, Shirouzu, M, Sekine, S, Kurumizaka, H. | | Deposit date: | 2018-06-25 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (8.7 Å) | | Cite: | Structural basis of the nucleosome transition during RNA polymerase II passage.
Science, 362, 2018
|
|
6A5T
 
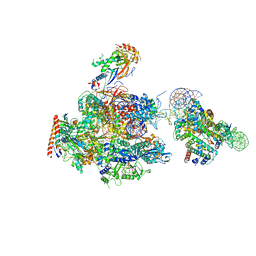 | | RNA polymerase II elongation complex stalled at SHL(-1) of the nucleosome | | Descriptor: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Kujirai, T, Ehara, H, Fujino, Y, Shirouzu, M, Sekine, S, Kurumizaka, H. | | Deposit date: | 2018-06-25 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Structural basis of the nucleosome transition during RNA polymerase II passage.
Science, 362, 2018
|
|
6A5P
 
 | | RNA polymerase II elongation complex stalled at SHL(-5) of the nucleosome | | Descriptor: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Kujirai, T, Ehara, H, Fujino, Y, Shirouzu, M, Sekine, S, Kurumizaka, H. | | Deposit date: | 2018-06-25 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Structural basis of the nucleosome transition during RNA polymerase II passage.
Science, 362, 2018
|
|
6A5U
 
 | | RNA polymerase II elongation complex stalled at SHL(-1) of the nucleosome, with foreign DNA, tilt conformation | | Descriptor: | DNA (198-MER), DNA (40-MER), DNA-directed RNA polymerase subunit, ... | | Authors: | Kujirai, T, Ehara, H, Fujino, Y, Shirouzu, M, Sekine, S, Kurumizaka, H. | | Deposit date: | 2018-06-25 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Structural basis of the nucleosome transition during RNA polymerase II passage.
Science, 362, 2018
|
|
6A5O
 
 | | RNA polymerase II elongation complex stalled at SHL(-6) of the nucleosome | | Descriptor: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Kujirai, T, Ehara, H, Fujino, Y, Shirouzu, M, Sekine, S, Kurumizaka, H. | | Deposit date: | 2018-06-25 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (9.9 Å) | | Cite: | Structural basis of the nucleosome transition during RNA polymerase II passage.
Science, 362, 2018
|
|
6INQ
 
 | | RNA polymerase II elongation complex stalled at SHL(-1) of the nucleosome, with foreign DNA (+1 position) | | Descriptor: | DNA (198-MER), DNA (31-MER), DNA-directed RNA polymerase subunit, ... | | Authors: | Kujirai, T, Ehara, H, Fujino, Y, Shirouzu, M, Sekine, S, Kurumizaka, H. | | Deposit date: | 2018-10-26 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Structural basis of the nucleosome transition during RNA polymerase II passage.
Science, 362, 2018
|
|
7C9J
 
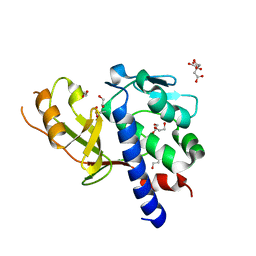 | | Transglutaminase from Geobacillus stearothermophilus (without C-terminal extension) | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, Protein-glutamine gamma-glutamyltransferase | | Authors: | Takita, T, Mikami, B, Lei, Y, Jing, Y, Yamada, A, Yasukawa, K. | | Deposit date: | 2020-06-06 | | Release date: | 2021-06-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Transglutaminase from Geobacillus stearothermophilus (without C-terminal extension)
To be published
|
|
6J50
 
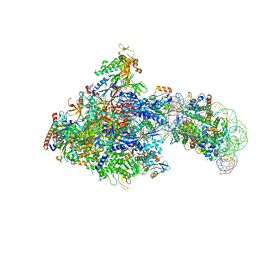 | | RNA polymerase II elongation complex bound with Spt4/5 and foreign DNA, stalled at SHL(-1) of the nucleosome (tilted conformation) | | Descriptor: | DNA (198-MER), DNA (41-MER), DNA-directed RNA polymerase subunit, ... | | Authors: | Ehara, H, Kujirai, T, Fujino, Y, Shirouzu, M, Kurumizaka, H, Sekine, S. | | Deposit date: | 2019-01-10 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structural insight into nucleosome transcription by RNA polymerase II with elongation factors.
Science, 363, 2019
|
|
6J4W
 
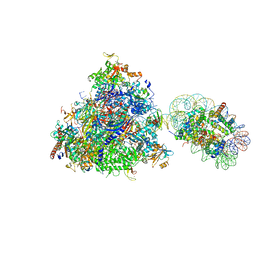 | | RNA polymerase II elongation complex bound with Elf1 and Spt4/5, stalled at SHL(-5) of the nucleosome | | Descriptor: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Ehara, H, Kujirai, T, Fujino, Y, Shirouzu, M, Kurumizaka, H, Sekine, S. | | Deposit date: | 2019-01-10 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Structural insight into nucleosome transcription by RNA polymerase II with elongation factors.
Science, 363, 2019
|
|
6J4X
 
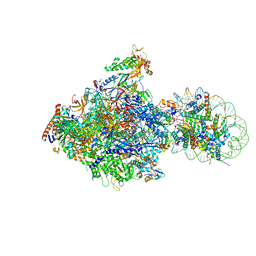 | | RNA polymerase II elongation complex bound with Elf1 and Spt4/5, stalled at SHL(-1) of the nucleosome (+1A) | | Descriptor: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Ehara, H, Kujirai, T, Fujino, Y, Shirouzu, M, Kurumizaka, H, Sekine, S. | | Deposit date: | 2019-01-10 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural insight into nucleosome transcription by RNA polymerase II with elongation factors.
Science, 363, 2019
|
|
