7DNY
 
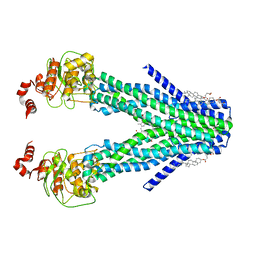 | | Cryo-EM structure of the human ABCB6 (coproporphyrin III-bound) | | Descriptor: | ATP-binding cassette sub-family B member 6, mitochondrial, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Kim, S, Lee, S.S, Park, J.G, Kim, J.W, Kim, S, Kim, N.J, Hong, S, Kang, J.Y, Jin, M.S. | | Deposit date: | 2020-12-11 | | Release date: | 2022-06-22 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural Insights into Porphyrin Recognition by the Human ATP-Binding Cassette Transporter ABCB6.
Mol.Cells, 45, 2022
|
|
5GW4
 
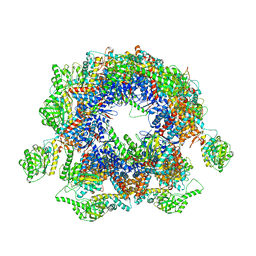 | | Structure of Yeast NPP-TRiC | | Descriptor: | T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, T-complex protein 1 subunit delta, ... | | Authors: | Zang, Y, Jin, M, Wang, H, Cong, Y. | | Deposit date: | 2016-09-08 | | Release date: | 2016-10-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Staggered ATP binding mechanism of eukaryotic chaperonin TRiC (CCT) revealed through high-resolution cryo-EM.
Nat. Struct. Mol. Biol., 23, 2016
|
|
5GW5
 
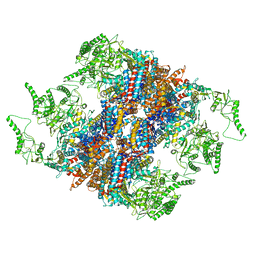 | | Structure of TRiC-AMP-PNP | | Descriptor: | T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, T-complex protein 1 subunit delta, ... | | Authors: | Zang, Y, Jin, M, Wang, H, Cong, Y. | | Deposit date: | 2016-09-08 | | Release date: | 2016-10-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Staggered ATP binding mechanism of eukaryotic chaperonin TRiC (CCT) revealed through high-resolution cryo-EM.
Nat. Struct. Mol. Biol., 23, 2016
|
|
1WWL
 
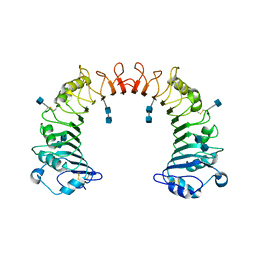 | | Crystal structure of CD14 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Monocyte differentiation antigen CD14 | | Authors: | Kim, J.-I, Lee, C.J, Jin, M.S, Lee, C.-H, Paik, S.-G, Lee, H, Lee, J.-O. | | Deposit date: | 2005-01-06 | | Release date: | 2005-02-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of CD14 and Its Implications for Lipopolysaccharide Signaling
J.Biol.Chem., 280, 2005
|
|
7CLT
 
 | | Crystal structure of the EFhd1/Swiprosin-2, a mitochondrial actin-binding protein | | Descriptor: | CALCIUM ION, EF-hand domain-containing protein D1, GLYCEROL, ... | | Authors: | Mun, S.A, Park, J, Park, K.R, Lee, Y, Kang, J.Y, Park, T, Jin, M, Yang, J, Jun, C.D, Eom, S.H. | | Deposit date: | 2020-07-22 | | Release date: | 2021-01-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07380986 Å) | | Cite: | Structural and Biochemical Characterization of EFhd1/Swiprosin-2, an Actin-Binding Protein in Mitochondria.
Front Cell Dev Biol, 8, 2020
|
|
4KNB
 
 | | C-Met in complex with OSI ligand | | Descriptor: | 7-[(1R)-1-(2,6-dichloro-3-fluorophenyl)ethoxy]-3-[1-(piperidin-4-yl)-1H-pyrazol-4-yl]furo[3,2-c]pyridin-6-amine, GAMMA-BUTYROLACTONE, Hepatocyte growth factor receptor | | Authors: | Wang, J, Steinig, A.G, Li, A.H, Chen, X, Dong, H, Ferraro, C, Jin, M, Kadalbajoo, M, Kleinberg, A, Stolz, K.M, Tavares-Greco, P.A, Wang, T, Albertella, M.R, Peng, Y, Crew, L, Kahler, J. | | Deposit date: | 2013-05-09 | | Release date: | 2014-04-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Novel 6-aminofuro[3,2-c]pyridines as potent, orally efficacious inhibitors of cMET and RON kinases.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4IP3
 
 | | Complex structure of OspI and Ubc13 | | Descriptor: | ORF169b, Ubiquitin-conjugating enzyme E2 N | | Authors: | Fu, P, Jin, M, Zhang, X, Xu, L, Xia, Z, Zhu, Y. | | Deposit date: | 2013-01-09 | | Release date: | 2013-03-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure Analysis of Ubc13 Inactivation
To be Published
|
|
7FZJ
 
 | | Crystal Structure of human FABP4 in complex with 3-cyclohexyl-2-ethylsulfanyl-6-hydroxypyrimidin-4-one | | Descriptor: | 1-cyclohexyl-2-(ethylsulfanyl)pyrimidine-4,6(1H,5H)-dione, FORMIC ACID, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Jin, M, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal Structure of a human FABP4 complex
To be published
|
|
6TUG
 
 | |
9EZY
 
 | | Vibrio cholerae DdmD-DdmE holo complex | | Descriptor: | 14 nucleotide DNA guide with terminal 5' phosphate, Helicase/UvrB N-terminal domain-containing protein, MAGNESIUM ION, ... | | Authors: | Loeff, L, Jinek, M. | | Deposit date: | 2024-04-14 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Molecular mechanism of plasmid elimination by the DdmDE defense system.
Science, 385, 2024
|
|
9EZX
 
 | | Vibrio cholerae DdmD apo complex | | Descriptor: | Helicase/UvrB N-terminal domain-containing protein | | Authors: | Loeff, L, Jinek, M. | | Deposit date: | 2024-04-14 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Molecular mechanism of plasmid elimination by the DdmDE defense system.
Science, 385, 2024
|
|
7Z4E
 
 | | SpCas9 bound to 8-nucleotide complementary DNA substrate | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, Non-target strand of 8 nucleotide complementary DNA substrate, Target strand of 8 nucleotide complementary DNA substrate, ... | | Authors: | Pacesa, M, Jinek, M. | | Deposit date: | 2022-03-03 | | Release date: | 2022-08-31 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.14 Å) | | Cite: | R-loop formation and conformational activation mechanisms of Cas9.
Nature, 609, 2022
|
|
7Z4I
 
 | | SpCas9 bound to 16-nucleotide complementary DNA substrate | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, Non-target strand of 16-nucleotide complementary DNA substrate, POTASSIUM ION, ... | | Authors: | Pacesa, M, Jinek, M. | | Deposit date: | 2022-03-03 | | Release date: | 2022-08-31 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | R-loop formation and conformational activation mechanisms of Cas9.
Nature, 609, 2022
|
|
7Z4H
 
 | | SpCas9 bound to 14-nucleotide complementary DNA substrate | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, Non-target strand of 14-nucleotide complementary DNA substrate, Target strand of 14-nucleotide complementary DNA substrate, ... | | Authors: | Pacesa, M, Jinek, M. | | Deposit date: | 2022-03-03 | | Release date: | 2022-08-31 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | R-loop formation and conformational activation mechanisms of Cas9.
Nature, 609, 2022
|
|
7Z4C
 
 | | SpCas9 bound to 6 nucleotide complementary DNA substrate | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, Non-target strand of 6 nucleotide complementary DNA substrate, Target strand of 6 nucleotide complementary DNA substrate, ... | | Authors: | Pacesa, M, Jinek, M. | | Deposit date: | 2022-03-03 | | Release date: | 2022-08-31 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | R-loop formation and conformational activation mechanisms of Cas9.
Nature, 609, 2022
|
|
7Z4G
 
 | | SpCas9 bound to 12-nucleotide complementary DNA substrate | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, Non-target strand of 12-nucleotide complementary DNA substrate, Target strand of 12-nucleotide complementary DNA substrate, ... | | Authors: | Pacesa, M, Jinek, M. | | Deposit date: | 2022-03-03 | | Release date: | 2022-08-31 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | R-loop formation and conformational activation mechanisms of Cas9.
Nature, 609, 2022
|
|
7Z4K
 
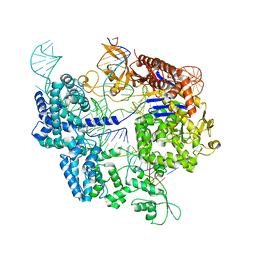 | | SpCas9 bound to 10-nucleotide complementary DNA substrate | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, Non-target strand of 10-nucleotide complementary DNA substrate, Target strand of 10-nucleotide complementary DNA substrate, ... | | Authors: | Pacesa, M, Jinek, M. | | Deposit date: | 2022-03-04 | | Release date: | 2022-08-31 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | R-loop formation and conformational activation mechanisms of Cas9.
Nature, 609, 2022
|
|
7Z4J
 
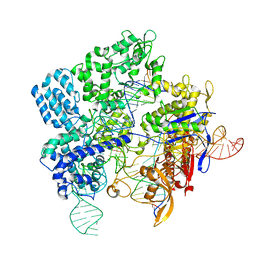 | |
7Z4L
 
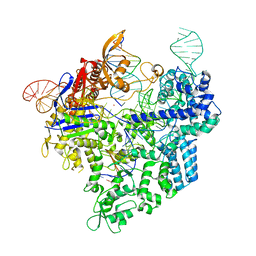 | |
7Z4D
 
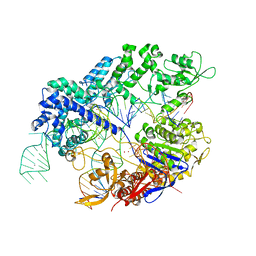 | |
4UN3
 
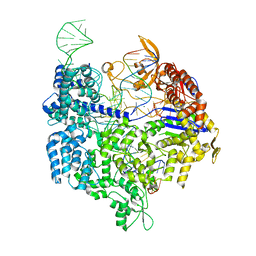 | | Crystal structure of Cas9 bound to PAM-containing DNA target | | Descriptor: | CRISPR-ASSOCIATED ENDONUCLEASE CAS9/CSN1, MAGNESIUM ION, NON-TARGET DNA STRAND, ... | | Authors: | Anders, C, Niewoehner, O, Duerst, A, Jinek, M. | | Deposit date: | 2014-05-25 | | Release date: | 2014-07-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.593 Å) | | Cite: | Structural Basis of Pam-Dependent Target DNA Recognition by the Cas9 Endonuclease
Nature, 513, 2014
|
|
4UN5
 
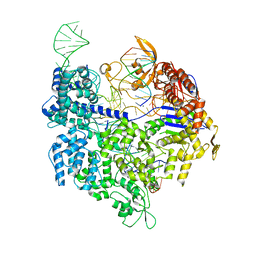 | | Crystal structure of Cas9 bound to PAM-containing DNA target containing mismatches at positions 1-3 | | Descriptor: | CRISPR-ASSOCIATED ENDONUCLEASE CAS9/CSN1, MAGNESIUM ION, NON-TARGET DNA STRAND, ... | | Authors: | Anders, C, Niewoehner, O, Duerst, A, Jinek, M. | | Deposit date: | 2014-05-25 | | Release date: | 2014-07-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of Pam-Dependent Target DNA Recognition by the Cas9 Endonuclease
Nature, 513, 2014
|
|
4UN4
 
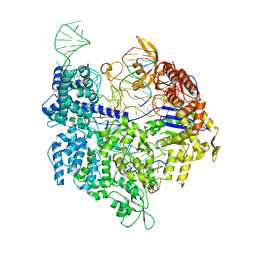 | | Crystal structure of Cas9 bound to PAM-containing DNA target with mismatches at positions 1-2 | | Descriptor: | CRISPR-ASSOCIATED ENDONUCLEASE CAS9/CSN1, MAGNESIUM ION, NON-TARGET DNA STRAND, ... | | Authors: | Anders, C, Niewoehner, O, Duerst, A, Jinek, M. | | Deposit date: | 2014-05-25 | | Release date: | 2014-07-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.371 Å) | | Cite: | Structural Basis of Pam-Dependent Target DNA Recognition by the Cas9 Endonuclease
Nature, 513, 2014
|
|
8QLP
 
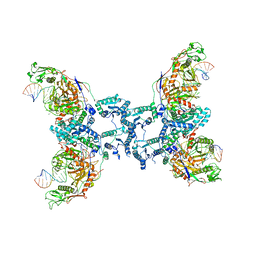 | | CryoEM structure of the RNA/DNA bound SPARTA (BabAgo/TIR-APAZ) tetrameric complex | | Descriptor: | DNA (5'-D(*AP*CP*TP*AP*AP*TP*AP*GP*AP*TP*TP*AP*GP*AP*GP*CP*CP*GP*TP*C)-3'), MAGNESIUM ION, RNA (5'-R(*AP*UP*GP*AP*CP*GP*GP*CP*UP*CP*UP*AP*AP*UP*CP*UP*AP*UP*UP*AP*GP*U)-3'), ... | | Authors: | Finocchio, G, Koopal, B, Potocnik, A, Heijstek, C, Jinek, M, Swarts, D. | | Deposit date: | 2023-09-20 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Target DNA-dependent activation mechanism of the prokaryotic immune system SPARTA.
Nucleic Acids Res., 52, 2024
|
|
7OX9
 
 | | Target-bound SpCas9 complex with AAVS1 all-RNA guide | | Descriptor: | AAVS1 non-target DNA strand, AAVS1 target DNA strand, CRISPR-associated endonuclease Cas9/Csn1, ... | | Authors: | Pacesa, M, Donohoue, P, May, A.P, Jinek, M, Cameron, P. | | Deposit date: | 2021-06-22 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Conformational control of Cas9 by CRISPR hybrid RNA-DNA guides mitigates off-target activity in T cells.
Mol.Cell, 81, 2021
|
|
