4JSC
 
 | | The 2.5A crystal structure of humanized Xenopus MDM2 with RO5316533 - a pyrrolidine MDM2 inhibitor | | Descriptor: | (3S,4R,5S)-3-(3-chloro-2-fluorophenyl)-4-(4-chloro-2-fluorophenyl)-4-cyano-N-[(3S)-3,4-dihydroxybutyl]-5-(2,2-dimethylpropyl)-D-prolinamide, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Janson, C.A, Lukacs, C, Graves, B. | | Deposit date: | 2013-03-22 | | Release date: | 2013-07-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of RG7388, a Potent and Selective p53-MDM2 Inhibitor in Clinical Development.
J.Med.Chem., 56, 2013
|
|
4J7D
 
 | | The 1.25A crystal structure of humanized Xenopus MDM2 with a nutlin fragment, RO5045331 | | Descriptor: | (4S,5R)-2-(4-tert-butyl-2-ethoxyphenyl)-4,5-bis(4-chlorophenyl)-4,5-dimethyl-4,5-dihydro-1H-imidazole, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Janson, C, Lukacs, C, Graves, B. | | Deposit date: | 2013-02-13 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Deconstruction of a nutlin: dissecting the binding determinants of a potent protein-protein interaction inhibitor.
ACS Med Chem Lett, 4, 2013
|
|
4J74
 
 | | The 1.2A crystal structure of humanized Xenopus MDM2 with RO0503918 - a nutlin fragment | | Descriptor: | (4S,5R)-4,5-bis(4-chlorophenyl)-2-methyl-4,5-dihydro-1H-imidazole, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Janson, C, Lukacs, C, Kammlott, U, Graves, B. | | Deposit date: | 2013-02-12 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Deconstruction of a nutlin: dissecting the binding determinants of a potent protein-protein interaction inhibitor.
ACS Med Chem Lett, 4, 2013
|
|
4J7E
 
 | | The 1.63A crystal structure of humanized Xenopus MDM2 with a nutlin fragment, RO5524529 | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, SULFATE ION, [(4S,5R)-4,5-bis(4-chlorophenyl)-2,4,5-trimethyl-4,5-dihydro-1H-imidazol-1-yl]{4-[3-(methylsulfonyl)propyl]piperazin-1-yl}methanone | | Authors: | Janson, C, Lukacs, C, Graves, B. | | Deposit date: | 2013-02-13 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Deconstruction of a nutlin: dissecting the binding determinants of a potent protein-protein interaction inhibitor.
ACS Med Chem Lett, 4, 2013
|
|
6HRP
 
 | |
1LX6
 
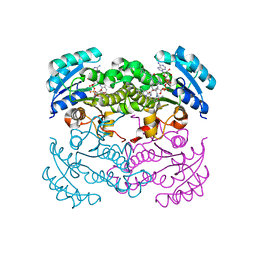 | |
1MFP
 
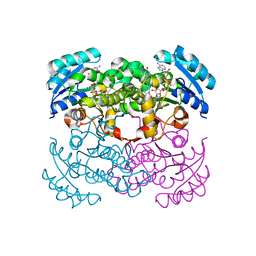 | | E. coli Enoyl Reductase in complex with NAD and SB611113 | | Descriptor: | (E)-N-METHYL-N-(1-METHYL-1H-INDOL-3-YLMETHYL)-3-(7-OXO-5,6,7,8-TETRAHYDRO-[1,8]NAPHTHYRIDIN-3-YL)-ACRYLAMIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Seefeld, M.A, Miller, W.H, Newlander, K.A, Burgess, W.J, DeWolf Jr, W.E, Elkins, P.A, Head, M.S, Jakas, D.R, Janson, C.A, Keller, P.M, Manley, P.J, Moore, T.D, Payne, D.J, Pearson, S, Polizzi, B.J, Qiu, X, Rittenhouse, S.F, Uzinskas, I.N, Wallis, N.G, Huffman, W.F. | | Deposit date: | 2002-08-13 | | Release date: | 2003-05-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Indole Naphthyridinones as Inhibitors of Bacterial Enoyl-ACP Reductases FabI and FabK
J.MED.CHEM., 46, 2003
|
|
3U15
 
 | | Structure of hDMX with Dimer Inducing Indolyl Hydantoin RO-2443 | | Descriptor: | (5Z)-5-[(6-chloro-7-methyl-1H-indol-3-yl)methylidene]-3-(3,4-difluorobenzyl)imidazolidine-2,4-dione, Protein Mdm4, SULFATE ION | | Authors: | Lukacs, C.M, Janson, C.A, Graves, B.J. | | Deposit date: | 2011-09-29 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Activation of the p53 pathway by small-molecule-induced MDM2 and MDMX dimerization.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2OAZ
 
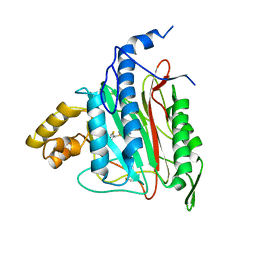 | | Human Methionine Aminopeptidase-2 Complexed with SB-587094 | | Descriptor: | COBALT (II) ION, N-(2-ISOPROPYLPHENYL)-3-[(2-THIENYLMETHYL)THIO]-1H-1,2,4-TRIAZOL-5-AMINE, human Methionine Amino Peptidase 2 | | Authors: | Marino Jr, J.P, Fisher, P.W, Hofmann, G.A, Kirkpatrick, R, Janson, C.A, Johnson, R.K, Ma, C, Mattern, M, Meek, T.D, Ryan, D, Schulz, C, Smith, W.W, Tew, D.G, Tomazek Jr, T.A, Veber, D.F, Xiong, W.C, Yamamoto, Y, Yamashita, K, Yang, G, Thompson, S.K. | | Deposit date: | 2006-12-18 | | Release date: | 2007-06-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Highly potent inhibitors of methionine aminopeptidase-2 based on a 1,2,4-triazole pharmacophore.
J.Med.Chem., 50, 2007
|
|
5C5Q
 
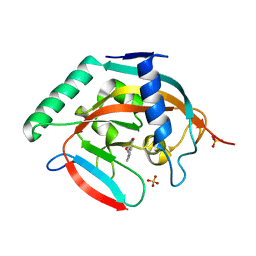 | | CRYSTAL STRUCTURE OF HUMAN TANKYRASE-2 IN COMPLEX WITH A PYRANOPYRIDONE INHIBITOR | | Descriptor: | (3R)-10-methyl-3-(propan-2-yl)-1,3,4,5-tetrahydro-6H-pyrano[4,3-c]isoquinolin-6-one, SULFATE ION, Tankyrase-2, ... | | Authors: | Lukacs, C.M, Janson, C.A. | | Deposit date: | 2015-06-21 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fragment-Based Drug Design of Novel Pyranopyridones as Cell Active and Orally Bioavailable Tankyrase Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
5C5P
 
 | | CRYSTAL STRUCTURE OF HUMAN TANKYRASE-2 IN COMPLEX WITH A PYRANOPYRIDONE INHIBITOR | | Descriptor: | (3R)-3-(1-hydroxy-2-methylpropan-2-yl)-1,3,4,5-tetrahydro-6H-pyrano[4,3-c]isoquinolin-6-one, SULFATE ION, Tankyrase-2, ... | | Authors: | Lukacs, C.M, Janson, C.A. | | Deposit date: | 2015-06-21 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Fragment-Based Drug Design of Novel Pyranopyridones as Cell Active and Orally Bioavailable Tankyrase Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
5C5R
 
 | | CRYSTAL STRUCTURE OF HUMAN TANKYRASE-2 IN COMPLEX WITH A PYRANOPYRIDONE INHIBITOR | | Descriptor: | (7R)-2-hydroxy-7-(propan-2-yl)-7,8-dihydro-5H-pyrano[4,3-b]pyridine-3-carbonitrile, SULFATE ION, Tankyrase-2, ... | | Authors: | Lukacs, C.M, Janson, C.A. | | Deposit date: | 2015-06-21 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Fragment-Based Drug Design of Novel Pyranopyridones as Cell Active and Orally Bioavailable Tankyrase Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
2ADU
 
 | | Human Methionine Aminopeptidase Complex with 4-Aryl-1,2,3-triazole Inhibitor | | Descriptor: | 4-(3-METHYLPHENYL)-1H-1,2,3-TRIAZOLE, COBALT (II) ION, Methionine aminopeptidase 2 | | Authors: | Kallander, L.S, Lu, Q, Chen, W, Tomaszek, T, Yang, G, Tew, D, Meek, T.D, Hofmann, G.A, Schulz-Pritchard, C.K, Smith, W.W, Janson, C.A, Ryan, M.D, Zhang, G.F, Johanson, K.O, Kirkpatrick, R.B, Ho, T.F, Fisher, P.W, Mattern, M.R, Johnson, R.K, Hansbury, M.J, Winkler, J.D, Ward, K.W, Veber, D.F, Thompson, S.K. | | Deposit date: | 2005-07-20 | | Release date: | 2005-09-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 4-Aryl-1,2,3-triazole: A Novel Template for a Reversible Methionine Aminopeptidase 2 Inhibitor, Optimized To Inhibit Angiogenesis in Vivo
J.Med.Chem., 48, 2005
|
|
8GJS
 
 | | Stapled Peptide ALRN-6924 Bound to MDMX | | Descriptor: | ACE-LEU-THR-PHE-ALA-GLU-TYR-TRP-ALA-GLN-LEU-DAL-ALA-ALA-ALA-ALA-ALA-DAL, Protein Mdm4 | | Authors: | Graves, B.J, Janson, C, Lukacs, C. | | Deposit date: | 2023-03-16 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of Sulanemadlin (ALRN-6924), the First Cell-Permeating, Stabilized alpha-Helical Peptide in Clinical Development.
J.Med.Chem., 66, 2023
|
|
1ZOW
 
 | | Crystal Structure of S. aureus FabH, beta-ketoacyl carrier protein synthase III | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase III | | Authors: | Qiu, X, Choudhry, A.E, Janson, C.A, Grooms, M, Daines, R.A, Lonsdale, J.T, Khandekar, S.S. | | Deposit date: | 2005-05-15 | | Release date: | 2005-08-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and substrate specificity of the beta-ketoacyl-acyl carrier protein synthase III (FabH) from Staphylococcus aureus.
Protein Sci., 14, 2005
|
|
3ORN
 
 | | Mitogen-activated protein kinase kinase 1 (MEK1) in complex with CH4987655 and MgAMP-PNP | | Descriptor: | 3,4-difluoro-2-[(2-fluoro-4-iodophenyl)amino]-N-(2-hydroxyethoxy)-5-[(3-oxo-1,2-oxazinan-2-yl)methyl]benzamide, Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, ... | | Authors: | Lukacs, C.M, Janson, C, Schuck, V, Belunis, C. | | Deposit date: | 2010-09-07 | | Release date: | 2011-03-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Design and synthesis of novel allosteric MEK inhibitor CH4987655 as an orally available anticancer agent.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3OS3
 
 | | Mitogen-activated protein kinase kinase 1 (MEK1) in complex with CH4858061 and MgATP | | Descriptor: | 2-[(4-ethynyl-2-fluorophenyl)amino]-3,4-difluoro-N-(2-hydroxyethoxy)-5-{[(2-hydroxyethoxy)imino]methyl}benzamide, ADENOSINE-5'-TRIPHOSPHATE, Dual specificity mitogen-activated protein kinase kinase 1, ... | | Authors: | Lukacs, C.M, Janson, C, Schuck, V, Belunis, C. | | Deposit date: | 2010-09-08 | | Release date: | 2011-07-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Design and synthesis of novel allosteric MEK inhibitor CH4987655 as an orally available anticancer agent.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
1NL6
 
 | |
1NLJ
 
 | |
1MZS
 
 | | CRYSTAL STRUCTURE OF BETA-KETOACYL-ACP SYNTHASE III WITH BOUND dichlorobenzyloxy-indole-carboxylic acid inhibitor | | Descriptor: | 1-(5-CARBOXYPENTYL)-5-(2,6-DICHLOROBENZYLOXY)-1H-INDOLE-2-CARBOXYLIC ACID, 3-oxoacyl-[acyl-carrier-protein] synthase III, PHOSPHATE ION | | Authors: | Daines, R.A, Pendrak, I, Sham, K, Van Aller, G.S, Konstantinidis, A.K, Lonsdale, J.T, Janson, C.A, Qui, X, Brandt, M, Silverman, C, Head, M.S. | | Deposit date: | 2002-10-09 | | Release date: | 2002-11-13 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | First X-ray cocrystal structure of a bacterial FabH condensing enzyme and a small molecule inhibitor achieved using rational design and homology modeling
J.Med.Chem., 46, 2003
|
|
4E73
 
 | | Crystal structure of JNK1beta-JIP in complex with an azaquinolone inhbitor | | Descriptor: | C-Jun-amino-terminal kinase-interacting protein 1, Mitogen-activated protein kinase 8, methyl 3-(4-{[(1R,2S,3S,5S,7s)-5-aminotricyclo[3.3.1.1~3,7~]dec-2-yl]carbamoyl}benzyl)-4-oxo-1-phenyl-1,4-dihydro-1,8-naphthyridine-2-carboxylate | | Authors: | Lukacs, C.M, Janson, C.A. | | Deposit date: | 2012-03-16 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Identification of an Adamantyl Azaquinolone JNK Selective Inhibitor.
ACS Med Chem Lett, 3, 2012
|
|
1GFW
 
 | |
1DDK
 
 | | CRYSTAL STRUCTURE OF IMP-1 METALLO BETA-LACTAMASE FROM PSEUDOMONAS AERUGINOSA | | Descriptor: | ACETIC ACID, IMP-1 METALLO BETA-LACTAMASE, ZINC ION | | Authors: | Concha, N.O, Janson, C.A, Rowling, P, Pearson, S, Cheever, C.A, Clarke, B.P, Lewis, C, Galleni, M, Frere, J.M, Payne, D.J, Bateson, J.H, Abdel-Meguid, S.S. | | Deposit date: | 1999-11-10 | | Release date: | 2000-11-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of the IMP-1 metallo beta-lactamase from Pseudomonas aeruginosa and its complex with a mercaptocarboxylate inhibitor: binding determinants of a potent, broad-spectrum inhibitor.
Biochemistry, 39, 2000
|
|
1DD6
 
 | | IMP-1 METALLO BETA-LACTAMASE FROM PSEUDOMONAS AERUGINOSA IN COMPLEX WITH A MERCAPTOCARBOXYLATE INHIBITOR | | Descriptor: | (2-MERCAPTOMETHYL-4-PHENYL-BUTYRYLIMINO)-(5-TETRAZOL-1-YLMETHYL-THIOPHEN-2-YL)-ACETIC ACID, IMP-1 METALLO BETA-LACTAMASE, SULFATE ION, ... | | Authors: | Concha, N.O, Janson, C.A, Rowling, P, Pearson, S, Cheever, C.A, Clarke, B.P, Lewis, C, Galleni, M, Frere, J.M, Payne, D.J, Bateson, J.H, Abdel-Meguid, S.S. | | Deposit date: | 1999-11-08 | | Release date: | 2000-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the IMP-1 metallo beta-lactamase from Pseudomonas aeruginosa and its complex with a mercaptocarboxylate inhibitor: binding determinants of a potent, broad-spectrum inhibitor.
Biochemistry, 39, 2000
|
|
1VJY
 
 | | Crystal Structure of a Naphthyridine Inhibitor of Human TGF-beta Type I Receptor | | Descriptor: | 2-[5-(6-METHYLPYRIDIN-2-YL)-2,3-DIHYDRO-1H-PYRAZOL-4-YL]-1,5-NAPHTHYRIDINE, TGF-beta receptor type I | | Authors: | Gellibert, F, Woolven, J, Fouchet, M.-H, Mathews, N, Goodland, H, Lovegrove, V, Laroze, A, Nguyen, V.-L, Sautet, S, Wang, R, Janson, C, Smith, W, Krysa, G, Boullay, V, de Gouville, A.-C, Huet, S, Hartley, D. | | Deposit date: | 2004-04-07 | | Release date: | 2004-08-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of 1,5-Naphthyridine Derivatives as a Novel Series of Potent and Selective TGF-beta Type I Receptor Inhibitors.
J.Med.Chem., 47, 2004
|
|
