5BK9
 
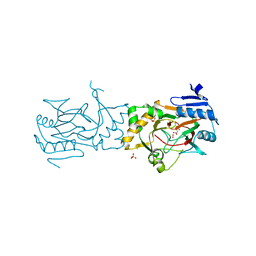 | | AAD-1 Bound to the Vanadyl Ion and Succinate | | Descriptor: | (R)-phenoxypropionate/alpha-ketoglutarate-dioxygenase, PHOSPHATE ION, SUCCINIC ACID, ... | | Authors: | Ongpipattanakul, C, Chekan, J.R. | | Deposit date: | 2019-06-01 | | Release date: | 2019-06-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Molecular basis for enantioselective herbicide degradation imparted by aryloxyalkanoate dioxygenases in transgenic plants.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6UHJ
 
 | | X-ray Structure of C148 mGFP | | Descriptor: | C148 mGFP | | Authors: | Winegar, P.W, Hayes, O.G, McMillan, J.R, Figg, C.A, Focia, P.J, Mirkin, C.A. | | Deposit date: | 2019-09-27 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | DNA-Directed Protein Packing within Single Crystals.
Chem, 6, 2020
|
|
4K7B
 
 | | Crystal structure of Extrinsic protein in photosystem II | | Descriptor: | Extrinsic protein in photosystem II, GLYCEROL, SULFATE ION | | Authors: | Nagao, R, Suga, M, Niikura, A, Okumura, A, Koua, F.H.M, Suzuki, T, Tomo, T, Enami, I, Shen, J.R. | | Deposit date: | 2013-04-16 | | Release date: | 2013-09-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of Psb31, a Novel Extrinsic Protein of Photosystem II from a Marine Centric Diatom and Implications for Its Binding and Function
Biochemistry, 52, 2013
|
|
6UHP
 
 | | Crystal Structure of C148 mGFP-ncDNA-1 | | Descriptor: | C148 mGFP-ncDNA-1 | | Authors: | Winegar, P.W, Hayes, O.G, McMillan, J.R, Figg, C.A, Focia, P.J, Mirkin, C.A. | | Deposit date: | 2019-09-27 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | DNA-Directed Protein Packing within Single Crystals.
Chem, 6, 2020
|
|
4ZPT
 
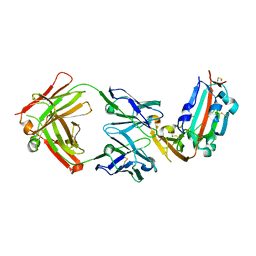 | | Structure of MERS-Coronavirus Spike Receptor-binding Domain (England1 Strain) in Complex with Vaccine-Elicited Murine Neutralizing Antibody D12 (Crystal Form 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D12 Fab Heavy chain, D12 Fab Light chain, ... | | Authors: | Joyce, M.G, Mascola, J.R, Graham, B.S, Kwong, P.D. | | Deposit date: | 2015-05-08 | | Release date: | 2015-10-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.591 Å) | | Cite: | Evaluation of candidate vaccine approaches for MERS-CoV
Nat Commun, 6, 2015
|
|
4GCB
 
 | |
5EZR
 
 | | Crystal Structure of PVX_084705 bound to compound | | Descriptor: | CHLORIDE ION, N-[5-(3-{2-[(cyclopropylmethyl)amino]pyrimidin-4-yl}-7-[(dimethylamino)methyl]-6-methylimidazo[1,2-a]pyridin-2-yl)-2-fluorophenyl]methanesulfonamide, cGMP-dependent protein kinase, ... | | Authors: | El Bakkouri, M, Amani, M, Walker, J.R, Osborne, S, Large, J.M, Birchall, K, Bouloc, N, Smiljanic-Hurley, E, Wheldon, M, Harding, D.J, Merritt, A.T, Ansell, K.H, Coombs, P.J, Kettleborough, C.A, Stewart, B.L, Bowyer, P.W, Gutteridge, W.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Baker, D.A, Hui, R, Loppnau, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-26 | | Release date: | 2017-05-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of PVX_084705 bound to compound
To Be Published
|
|
6UK4
 
 | |
6UKI
 
 | |
6U07
 
 | | Computational Stabilization of T Cell Receptor Constant Domains | | Descriptor: | MAGNESIUM ION, Stabilized T cell receptor constant domain (Calpha), Stabilized T cell receptor constant domain (Cbeta) | | Authors: | Froning, K, Maguire, J, Sereno, A, Huang, F, Chang, S, Weichert, K, Frommelt, A.J, Dong, J, Wu, X, Austin, H, Conner, E.M, Fitchett, J.R, Heng, A.R, Balasubramaniam, D, Hilgers, M.T, Kuhlman, B, Demarest, S.J. | | Deposit date: | 2019-08-13 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Computational stabilization of T cell receptors allows pairing with antibodies to form bispecifics.
Nat Commun, 11, 2020
|
|
5B82
 
 | |
5BKB
 
 | | Crystal structure of AAD-1 in complex with (R)-dichlorprop, Mn(II), and 2-oxoglutarate | | Descriptor: | (2R)-2-(2,4-dichlorophenoxy)propanoic acid, (R)-phenoxypropionate/alpha-ketoglutarate-dioxygenase, 2-OXOGLUTARIC ACID, ... | | Authors: | Chekan, J.R, Nair, S.K. | | Deposit date: | 2019-06-02 | | Release date: | 2019-06-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.582 Å) | | Cite: | Molecular basis for enantioselective herbicide degradation imparted by aryloxyalkanoate dioxygenases in transgenic plants.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
1BLS
 
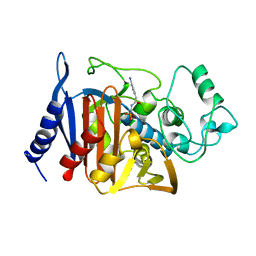 | |
4G2P
 
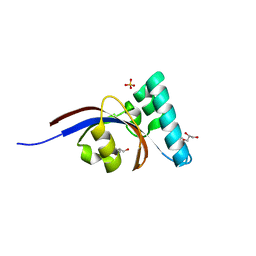 | | Crystal structure of peptidyl-prolyl cis-trans isomerase domain II of molecular chaperone SurA from Salmonella enterica subsp. enterica serovar Typhimurium str. 14028S | | Descriptor: | Chaperone SurA, GLYCEROL, SULFATE ION | | Authors: | Chang, C, Wu, R, Adkins, J.N, Brown, R.N, Cort, J.R, Heffron, F, Nakayasu, E.S, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Program for the Characterization of Secreted Effector Proteins (PCSEP) | | Deposit date: | 2012-07-12 | | Release date: | 2012-08-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of peptidyl-prolyl cis-trans isomerase domain II of molecular chaperone SurA from Salmonella enterica subsp. enterica serovar Typhimurium str. 14028S
TO BE PUBLISHED
|
|
6UHL
 
 | | Crystal Structure of C148 mGFP-scDNA-1 | | Descriptor: | C148 mGFP-scDNA-1, UNKNOWN LIGAND | | Authors: | Winegar, P.W, Hayes, O.G, McMillan, J.R, Figg, C.A, Focia, P.J, Mirkin, C.A. | | Deposit date: | 2019-09-27 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | DNA-Directed Protein Packing within Single Crystals.
Chem, 6, 2020
|
|
4G4C
 
 | | Room temperature X-ray diffraction study of carboplatin binding to HEWL in DMSO media after 13 months of crystal storage | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Lysozyme C, ... | | Authors: | Tanley, S.W.M, Schreurs, A.M.M, Kroon-Batenburg, L.M.J, Helliwell, J.R. | | Deposit date: | 2012-07-16 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Room-temperature X-ray diffraction studies of cisplatin and carboplatin binding to His15 of HEWL after prolonged chemical exposure.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
6UKE
 
 | |
5BQM
 
 | | Crystal structure of SXN101959, a Clostridium botulinum neurotoxin type D derivative and targeted secretion inhibitor | | Descriptor: | Botulinum neurotoxin type D, Somatoliberin,Botulinum neurotoxin type D, ZINC ION | | Authors: | Masuyer, G, Davies, J.R, Moore, K, Chaddock, J.A, Acharya, K.R. | | Deposit date: | 2015-05-29 | | Release date: | 2015-08-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural analysis of Clostridium botulinum neurotoxin type D as a platform for the development of targeted secretion inhibitors.
Sci Rep, 5, 2015
|
|
4GCF
 
 | |
6UHM
 
 | | Crystal Structure of a Physical Mixture of C148 mGFP and scDNA-1 | | Descriptor: | C148 mGFP | | Authors: | Winegar, P.W, Hayes, O.G, McMillan, J.R, Figg, C.A, Focia, P.J, Mirkin, C.A. | | Deposit date: | 2019-09-27 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | DNA-Directed Protein Packing within Single Crystals.
Chem, 6, 2020
|
|
5F0A
 
 | | CRYSTAL STRUCTURE OF PVX_084705 WITH BOUND 1-tert-butyl-3-(3-chlorophenoxy)-1H-pyrazolo[3,4-d]pyrimidin-4-amine INHIBITOR | | Descriptor: | 1-tert-butyl-3-(3-chlorophenoxy)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, cGMP-dependent protein kinase, putative | | Authors: | Walker, J.R, Wernimont, A.K, He, H, Seitova, A, Loppnau, P, Sibley, L.D, Graslund, S, Hutchinson, A, Bountra, C, Weigelt, J, Edwards, A.M, Arrowsmith, C.H, Hui, R, El Bakkouri, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-27 | | Release date: | 2015-12-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | CRYSTAL STRUCTURE OF PVX_084705 WITH BOUND INHIBITOR
To be published
|
|
4GCC
 
 | |
6UJQ
 
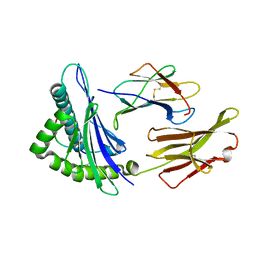 | |
5EYV
 
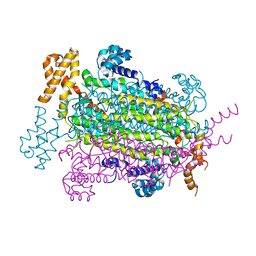 | | Crystal Structure of Adenylosuccinate lyase from Schistosoma mansoni in APO form. | | Descriptor: | Adenylosuccinate lyase | | Authors: | Romanello, L, Torini, J.R, Bird, L.E, Nettleship, J.E, Owens, R.J, Reddivari, Y, Brandao-Neto, J, Pereira, H.M. | | Deposit date: | 2015-11-25 | | Release date: | 2016-11-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structural and kinetic analysis of Schistosoma mansoni Adenylosuccinate Lyase (SmADSL).
Mol. Biochem. Parasitol., 214, 2017
|
|
4L6I
 
 | | Methylthioadenosine phosphorylase from Schistosoma mansoni in complex with adenine | | Descriptor: | ADENINE, S-methyl-5'-thioadenosine phosphorylase, SULFATE ION | | Authors: | Torini, J.R, DeMarco, R, Brandao-Neto, J, Pereira, H.M. | | Deposit date: | 2013-06-12 | | Release date: | 2014-06-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Schistosoma mansoni Adenosine Phosphorylase/5'-Methylthioadenosine Phosphorylase and Its Importance on Adenosine Salvage Pathway.
Plos Negl Trop Dis, 10, 2016
|
|
