6I2H
 
 | |
1DGJ
 
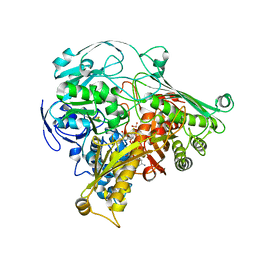 | | CRYSTAL STRUCTURE OF THE ALDEHYDE OXIDOREDUCTASE FROM DESULFOVIBRIO DESULFURICANS ATCC 27774 | | Descriptor: | ALDEHYDE OXIDOREDUCTASE, FE2/S2 (INORGANIC) CLUSTER, MOLYBDENUM (IV)OXIDE, ... | | Authors: | Rebelo, J.M, Macieira, S, Dias, J.M, Huber, R, Romao, M.J. | | Deposit date: | 1999-11-24 | | Release date: | 2000-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Gene sequence and crystal structure of the aldehyde oxidoreductase from Desulfovibrio desulfuricans ATCC 27774.
J.Mol.Biol., 297, 2000
|
|
6ZTR
 
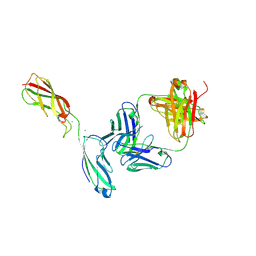 | | Crystal Structure of the anti-human P-Cadherin Fab CQY684 in complex with human P-Cadherin(108-324) | | Descriptor: | CALCIUM ION, CQY684 Fab heavy-chain, CQY684 Fab light-chain, ... | | Authors: | Rondeau, J.M, Lehmann, S. | | Deposit date: | 2020-07-20 | | Release date: | 2021-05-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | PCA062, a P-cadherin Targeting Antibody-Drug Conjugate, Displays Potent Antitumor Activity Against P-cadherin-expressing Malignancies.
Mol.Cancer Ther., 20, 2021
|
|
6ZTB
 
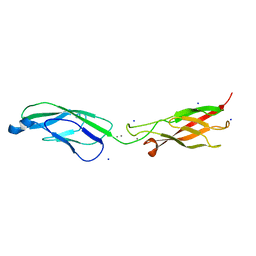 | | Crystal Structure of human P-Cadherin EC1_EC2 | | Descriptor: | CALCIUM ION, Cadherin-3, SODIUM ION | | Authors: | Rondeau, J.M, Lehmann, S. | | Deposit date: | 2020-07-17 | | Release date: | 2021-05-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | PCA062, a P-cadherin Targeting Antibody-Drug Conjugate, Displays Potent Antitumor Activity Against P-cadherin-expressing Malignancies.
Mol.Cancer Ther., 20, 2021
|
|
6ZTF
 
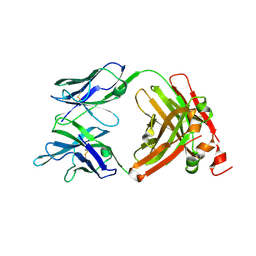 | | Crystal Structure of the anti-human P-Cadherin Fab CQY684 | | Descriptor: | CQY684 Fab heavy-chain, CQY684 Fab light-chain | | Authors: | Rondeau, J.M, Lehmann, S. | | Deposit date: | 2020-07-20 | | Release date: | 2021-05-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | PCA062, a P-cadherin Targeting Antibody-Drug Conjugate, Displays Potent Antitumor Activity Against P-cadherin-expressing Malignancies.
Mol.Cancer Ther., 20, 2021
|
|
6IC4
 
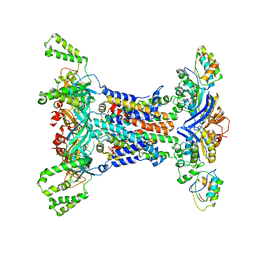 | |
6I7M
 
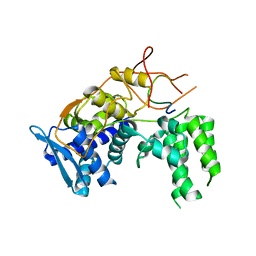 | | Influenza A nucleoprotein docked into 3D helical structure of the wild type ribonucleoprotein complex obtained using cryoEM. Conformation 4. | | Descriptor: | Nucleoprotein | | Authors: | Coloma, R, Arranz, R, de la Rosa-Trevin, J.M, Sorzano, C.O.S, Carlero, D, Ortin, J, Martin-Benito, J. | | Deposit date: | 2018-11-16 | | Release date: | 2020-02-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Structural insights into influenza A virus ribonucleoproteins reveal a processive helical track as transcription mechanism.
Nat Microbiol, 5, 2020
|
|
6IA2
 
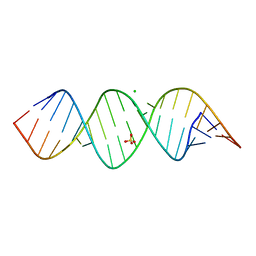 | | Crystal structure of a self-complementary RNA duplex recognized by Com | | Descriptor: | CHLORIDE ION, RNA (5'-R(*AP*GP*AP*GP*AP*AP*CP*CP*CP*GP*GP*AP*GP*UP*UP*CP*CP*CP*U)-3'), SULFATE ION | | Authors: | Nowacka, M, Fernandes, H, Kiliszek, A, Bernat, A, Lach, G, Bujnicki, J.M. | | Deposit date: | 2018-11-26 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Specific interaction of zinc finger protein Com with RNA and the crystal structure of a self-complementary RNA duplex recognized by Com.
Plos One, 14, 2019
|
|
6I2A
 
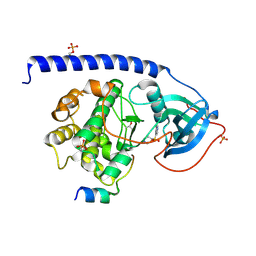 | | Crystal Structure of the Protein-Kinase A catalytic subunit from Cricetulus Griseus in complex with compounds RKp153 and Fasudil | | Descriptor: | 5-(1,4-DIAZEPAN-1-SULFONYL)ISOQUINOLINE, UPF0418 protein FAM164A, beta-D-ribopyranose, ... | | Authors: | Mueller, J.M, Heine, A, Klebe, G. | | Deposit date: | 2018-11-01 | | Release date: | 2019-05-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Conceptional Design of Self-Assembling Bisubstrate-like Inhibitors of Protein Kinase A Resulting in a Boronic Acid Glutamate Linkage
Acs Omega, 2019
|
|
6I7B
 
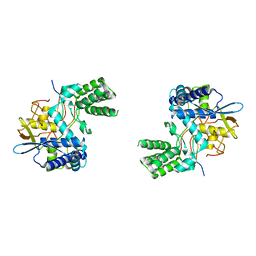 | | Influenza A nucleoprotein docked into 3D helical structure of the wild type ribonucleoprotein complex obtained using cryoEM. Conformation 3. | | Descriptor: | Nucleoprotein | | Authors: | Coloma, R, Arranz, R, de la Rosa-Trevin, J.M, Sorzano, C.O.S, Carlero, D, Ortin, J, Martin-Benito, J. | | Deposit date: | 2018-11-16 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Structural insights into influenza A virus ribonucleoproteins reveal a processive helical track as transcription mechanism.
Nat Microbiol, 5, 2020
|
|
6ZVP
 
 | | Atomic model of the EM-based structure of the full-length tyrosine hydroxylase in complex with dopamine (residues 40-497) in which the regulatory domain (residues 40-165) has been included only with the backbone atoms | | Descriptor: | FE (III) ION, L-DOPAMINE, Tyrosine 3-monooxygenase | | Authors: | Bueno-Carrasco, M.T, Cuellar, J, Santiago, C, Valpuesta, J.M, Martinez, A, Flydal, M.I. | | Deposit date: | 2020-07-27 | | Release date: | 2021-11-17 | | Last modified: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural mechanism for tyrosine hydroxylase inhibition by dopamine and reactivation by Ser40 phosphorylation.
Nat Commun, 13, 2022
|
|
7TQV
 
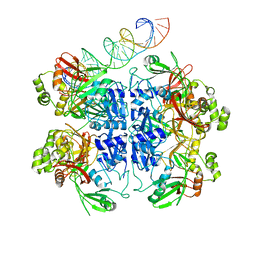 | | SARS-CoV-2 endoribonuclease Nsp15 bound to dsRNA | | Descriptor: | RNA (33-MER), Uridylate-specific endoribonuclease | | Authors: | Frazier, M.N, Krahn, J.M, Butay, K.J, Dillard, L.B, Borgnia, M.J, Stanley, R.E. | | Deposit date: | 2022-01-27 | | Release date: | 2022-03-23 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Flipped over U: structural basis for dsRNA cleavage by the SARS-CoV-2 endoribonuclease.
Nucleic Acids Res., 50, 2022
|
|
6ZZU
 
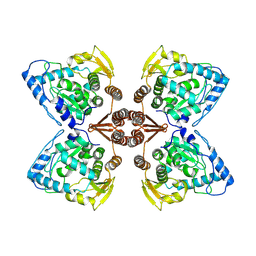 | | Partial structure of the substrate-free tyrosine hydroxylase (apo-TH). | | Descriptor: | FE (III) ION, Tyrosine 3-monooxygenase | | Authors: | Bueno-Carrasco, M.T, Cuellar, J, Santiago, C, Valpuesta, J.M, Martinez, A, Flydal, M.I. | | Deposit date: | 2020-08-05 | | Release date: | 2021-11-17 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural mechanism for tyrosine hydroxylase inhibition by dopamine and reactivation by Ser40 phosphorylation.
Nat Commun, 13, 2022
|
|
7A2G
 
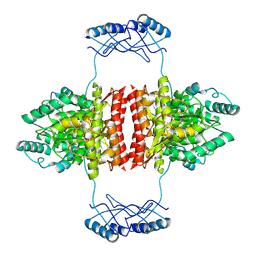 | | Full-length structure of the substrate-free tyrosine hydroxylase (apo-TH). | | Descriptor: | FE (III) ION, Tyrosine 3-monooxygenase | | Authors: | Bueno-Carrasco, M.T, Cuellar, J, Santiago, C, Flydal, M.I, Martinez, A, Valpuesta, J.M. | | Deposit date: | 2020-08-17 | | Release date: | 2021-12-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural mechanism for tyrosine hydroxylase inhibition by dopamine and reactivation by Ser40 phosphorylation.
Nat Commun, 13, 2022
|
|
6ZN2
 
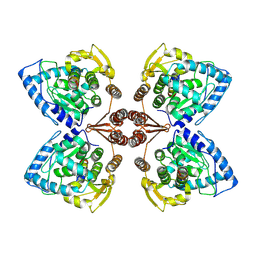 | | Partial structure of tyrosine hydroxylase in complex with dopamine showing the catalytic domain and an alpha-helix from the regulatory domain involved in dopamine binding. | | Descriptor: | FE (III) ION, L-DOPAMINE, SER-LEU-ILE-GLU-ASP-ALA-ARG-LYS-GLU-ARG-GLU-ALA-ALA-VAL-ALA-ALA-ALA-ALA, ... | | Authors: | Bueno-Carrasco, M.T, Cuellar, J, Santiago, C, Valpuesta, J.M, Martinez, A, Flydal, M.I. | | Deposit date: | 2020-07-06 | | Release date: | 2021-12-08 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural mechanism for tyrosine hydroxylase inhibition by dopamine and reactivation by Ser40 phosphorylation.
Nat Commun, 13, 2022
|
|
7TJ2
 
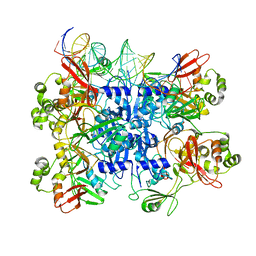 | | SARS-CoV-2 endoribonuclease Nsp15 bound to dsRNA | | Descriptor: | RNA (31-MER), Uridylate-specific endoribonuclease nsp15 | | Authors: | Frazier, M.N, Krahn, J.M, Butay, K.J, Dillard, L.B, Borgnia, M.J, Stanley, R.E. | | Deposit date: | 2022-01-14 | | Release date: | 2022-03-23 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Flipped over U: structural basis for dsRNA cleavage by the SARS-CoV-2 endoribonuclease.
Nucleic Acids Res., 50, 2022
|
|
7AAL
 
 | | Crystal structure of the F-BAR domain of PSTIPIP1, G258A mutant | | Descriptor: | Proline-serine-threonine phosphatase-interacting protein 1 | | Authors: | Manso, J.A, Alcon, P, Bayon, Y, Alonso, A, de Pereda, J.M. | | Deposit date: | 2020-09-04 | | Release date: | 2022-02-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | PSTPIP1-LYP phosphatase interaction: structural basis and implications for autoinflammatory disorders.
Cell.Mol.Life Sci., 79, 2022
|
|
7AAM
 
 | | Crystal structure of the F-BAR domain of PSTIPIP1 bound to the CTH domain of the phosphatase LYP | | Descriptor: | GLYCEROL, Proline-serine-threonine phosphatase-interacting protein 1, Tyrosine-protein phosphatase non-receptor type 22 | | Authors: | Manso, J.A, Alcon, P, Bayon, Y, Alonso, A, de Pereda, J.M. | | Deposit date: | 2020-09-04 | | Release date: | 2022-02-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | PSTPIP1-LYP phosphatase interaction: structural basis and implications for autoinflammatory disorders.
Cell.Mol.Life Sci., 79, 2022
|
|
7AAN
 
 | | Crystal structure of the F-BAR domain of PSTIPIP1 | | Descriptor: | Proline-serine-threonine phosphatase-interacting protein 1 | | Authors: | Manso, J.A, Alcon, P, Bayon, Y, Alonso, A, de Pereda, J.M. | | Deposit date: | 2020-09-04 | | Release date: | 2022-02-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | PSTPIP1-LYP phosphatase interaction: structural basis and implications for autoinflammatory disorders.
Cell.Mol.Life Sci., 79, 2022
|
|
6J5W
 
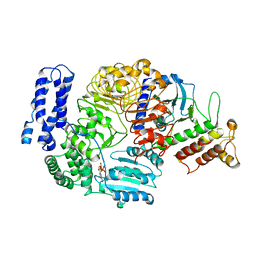 | | Ligand-triggered allosteric ADP release primes a plant NLR complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Disease resistance RPP13-like protein 4, RKS1 | | Authors: | Wang, J.Z, Wang, J, Hu, M.J, Wang, H.W, Zhou, J.M, Chai, J.J. | | Deposit date: | 2019-01-12 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Ligand-triggered allosteric ADP release primes a plant NLR complex.
Science, 364, 2019
|
|
5DGS
 
 | |
6UX8
 
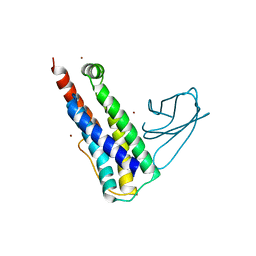 | |
6J6I
 
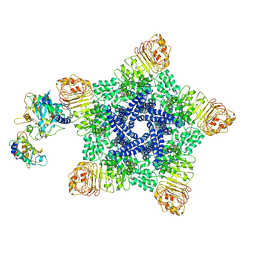 | | Reconstitution and structure of a plant NLR resistosome conferring immunity | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Disease resistance RPP13-like protein 4, Probable serine/threonine-protein kinase PBL2, ... | | Authors: | Wang, J.Z, Wang, J, Hu, M.J, Wang, H.W, Zhou, J.M, Chai, J.J. | | Deposit date: | 2019-01-15 | | Release date: | 2019-03-20 | | Last modified: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Reconstitution and structure of a plant NLR resistosome conferring immunity.
Science, 364, 2019
|
|
6J5T
 
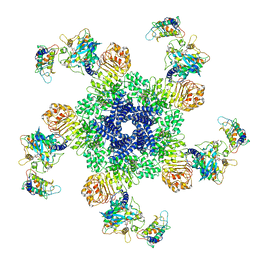 | | Reconstitution and structure of a plant NLR resistosome conferring immunity | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Disease resistance RPP13-like protein 4, Probable serine/threonine-protein kinase PBL2, ... | | Authors: | Wang, J.Z, Wang, J, Hu, M.J, Wang, H.W, Zhou, J.M, Chai, J.J. | | Deposit date: | 2019-01-12 | | Release date: | 2019-03-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Reconstitution and structure of a plant NLR resistosome conferring immunity.
Science, 364, 2019
|
|
7AQO
 
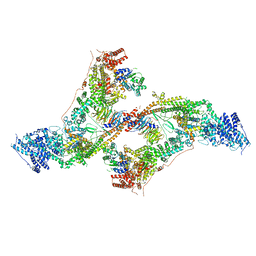 | | yeast THO-Sub2 complex dimer | | Descriptor: | BJ4_G0025130.mRNA.1.CDS.1, EM14S01-3B_G0007820.mRNA.1.CDS.1, TEX1 isoform 1, ... | | Authors: | Schuller, S.K, Schuller, J.M, Prabu, R.J, Baumgartner, M, Bonneau, F, basquin, J, Conti, E. | | Deposit date: | 2020-10-22 | | Release date: | 2020-12-02 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural insights into the nucleic acid remodeling mechanisms of the yeast THO-Sub2 complex.
Elife, 9, 2020
|
|
