6P7M
 
 | | Cryo-EM structure of LbCas12a-crRNA: AcrVA4 (1:2 complex) | | Descriptor: | Cas12a, MAGNESIUM ION, anti-CRISPR VA4, ... | | Authors: | Knott, G.J, Liu, J.J, Doudna, J.A. | | Deposit date: | 2019-06-06 | | Release date: | 2019-08-21 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for AcrVA4 inhibition of specific CRISPR-Cas12a.
Elife, 8, 2019
|
|
7ZDD
 
 | | Crystal structure of TRIM33 PHD-Bromodomain isoform B in complex with H3K10ac histone peptide. | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase TRIM33, Histone H3.X, ... | | Authors: | Caria, S, Duclos, S, Crespillo, S, Errey, J, Barker, J.J. | | Deposit date: | 2022-03-29 | | Release date: | 2022-06-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.625 Å) | | Cite: | Identification of Histone Peptide Binding Specificity and Small-Molecule Ligands for the TRIM33 alpha and TRIM33 beta Bromodomains.
Acs Chem.Biol., 17, 2022
|
|
6FRH
 
 | | Crystal structure of Ssp DnaB Mini-Intein variant M86 | | Descriptor: | Replicative DNA helicase,Replicative DNA helicase | | Authors: | Popp, M.A, Blankenfeldt, W, Gazdag, M.E, Matern, J.J.C, Mootz, H.D. | | Deposit date: | 2018-02-15 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | A functional interplay between intein and extein sequences in protein splicing compensates for the essential block B histidine.
Chem Sci, 10, 2019
|
|
6P7N
 
 | | Cryo-EM structure of LbCas12a-crRNA: AcrVA4 (2:2 complex) | | Descriptor: | Cas12a, MAGNESIUM ION, anti-CRISPR VA4, ... | | Authors: | Knott, G.J, Liu, J.J, Doudna, J.A. | | Deposit date: | 2019-06-06 | | Release date: | 2019-08-21 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structural basis for AcrVA4 inhibition of specific CRISPR-Cas12a.
Elife, 8, 2019
|
|
7YOJ
 
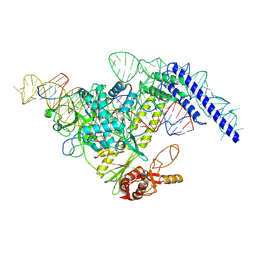 | | Structure of CasPi with guide RNA and target DNA | | Descriptor: | CasPi, DNA (30-MER), DNA (5'-D(P*CP*GP*GP*GP*AP*TP*GP*CP*CP*CP*AP*G)-3'), ... | | Authors: | Li, C.P, Wang, J, Liu, J.J. | | Deposit date: | 2022-08-01 | | Release date: | 2023-02-15 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | The compact Cas pi (Cas12l) 'bracelet' provides a unique structural platform for DNA manipulation.
Cell Res., 33, 2023
|
|
6QBT
 
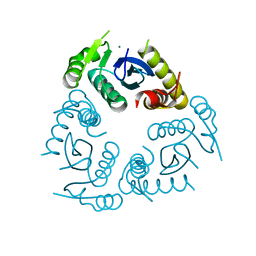 | |
6FPT
 
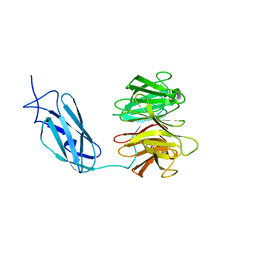 | | Crystal structure of Danio rerio Lin41 filamin-NHL domains | | Descriptor: | E3 ubiquitin-protein ligase TRIM71 | | Authors: | Kumari, P, Aeschimann, F, Gaidatzis, D, Keusch, J.J, Ghosh, P, Neagu, A, Pachulska-Wieczorek, K, Bujnicki, J.M, Gut, H, Grosshans, H, Ciosk, R. | | Deposit date: | 2018-02-12 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Evolutionary plasticity of the NHL domain underlies distinct solutions to RNA recognition.
Nat Commun, 9, 2018
|
|
6FQL
 
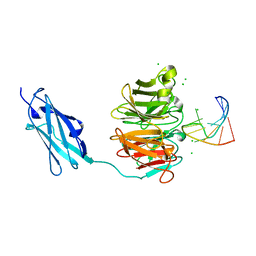 | | Crystal structure of Danio rerio Lin41 filamin-NHL domains in complex with mab-10 3'UTR 13mer RNA | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase TRIM71, RNA (5'-R(*UP*GP*CP*AP*UP*UP*UP*AP*AP*UP*GP*CP*A)-3') | | Authors: | Kumari, P, Aeschimann, F, Gaidatzis, D, Keusch, J.J, Ghosh, P, Neagu, A, Pachulska-Wieczorek, K, Bujnicki, J.M, Gut, H, Grosshans, H, Ciosk, R. | | Deposit date: | 2018-02-14 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.349 Å) | | Cite: | Evolutionary plasticity of the NHL domain underlies distinct solutions to RNA recognition.
Nat Commun, 9, 2018
|
|
6QJO
 
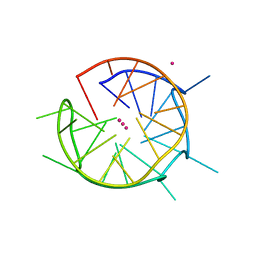 | | DNA containing both right- and left-handed parallel-stranded G-quadruplexes | | Descriptor: | DNA (28-MER), POTASSIUM ION | | Authors: | Winnerdy, F.R, Bakalar, B, Maity, A, Vandana, J.J, Schmitt, E, Mechulam, Y, Phan, A.T. | | Deposit date: | 2019-01-24 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | NMR solution and X-ray crystal structures of a DNA molecule containing both right- and left-handed parallel-stranded G-quadruplexes.
Nucleic Acids Res., 47, 2019
|
|
6PU1
 
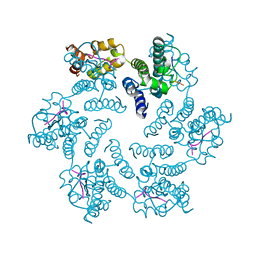 | |
6GCI
 
 | | Structure of the bongkrekic acid-inhibited mitochondrial ADP/ATP carrier | | Descriptor: | Bongkrekic acid, CARDIOLIPIN, HEXAETHYLENE GLYCOL, ... | | Authors: | Ruprecht, J.J, King, M.S, Pardon, E, Aleksandrova, A.A, Zogg, T, Crichton, P.G, Steyaert, J, Kunji, E.R.S. | | Deposit date: | 2018-04-17 | | Release date: | 2019-01-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The Molecular Mechanism of Transport by the Mitochondrial ADP/ATP Carrier.
Cell, 176, 2019
|
|
6QK2
 
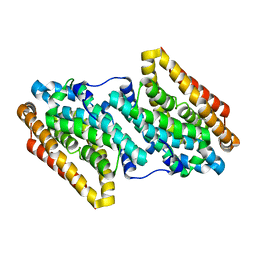 | |
6GV4
 
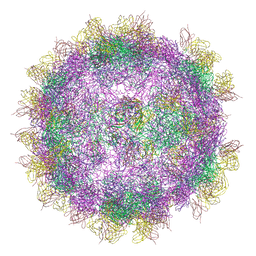 | | High-resolution Cryo-EM of Fab-labeled human parechovirus 3 | | Descriptor: | AT12-015 antibody variable heavy, AT12-015 antibody variable light, RNA (5'-R(*UP*GP*GP*UP*AP*UP*UP*U)-3'), ... | | Authors: | Domanska, A, Flatt, J.W, Jukonen, J.J.J, Geraets, J.A, Butcher, S.J. | | Deposit date: | 2018-06-20 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | A 2.8-Angstrom-Resolution Cryo-Electron Microscopy Structure of Human Parechovirus 3 in Complex with Fab from a Neutralizing Antibody.
J.Virol., 93, 2019
|
|
6QK0
 
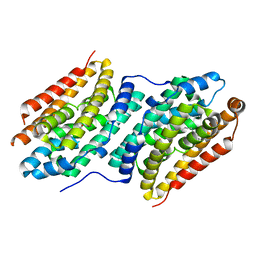 | |
8A64
 
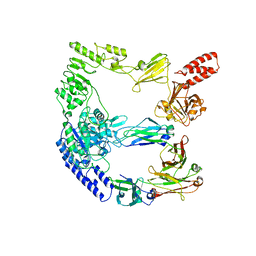 | | cryoEM structure of the catalytically inactive EndoS from S. pyogenes in complex with the Fc region of immunoglobulin G1. | | Descriptor: | Endo-beta-N-acetylglucosaminidase F2, Immunoglobulin gamma-1 heavy chain, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Trastoy, B, Cifuente, J.O, Du, J.J, Sundberg, E.J, Guerin, M.E. | | Deposit date: | 2022-06-16 | | Release date: | 2023-04-05 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Mechanism of antibody-specific deglycosylation and immune evasion by Streptococcal IgG-specific endoglycosidases.
Nat Commun, 14, 2023
|
|
6O4D
 
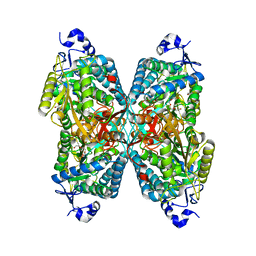 | | Structure of ALDH7A1 mutant W175A complexed with L-pipecolic acid | | Descriptor: | 1,2-ETHANEDIOL, Alpha-aminoadipic semialdehyde dehydrogenase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tanner, J.J, Korasick, D.A, Laciak, A.R. | | Deposit date: | 2019-02-28 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural and biochemical consequences of pyridoxine-dependent epilepsy mutations that target the aldehyde binding site of aldehyde dehydrogenase ALDH7A1.
Febs J., 287, 2020
|
|
6ODC
 
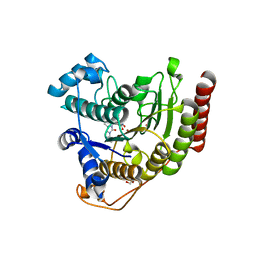 | | Crystal structure of HDAC8 in complex with compound 30 | | Descriptor: | (2E)-3-[2-(3-cyclopentyl-5,5-dimethyl-2-oxoimidazolidin-1-yl)phenyl]-N-hydroxyprop-2-enamide, 1,2-ETHANEDIOL, Histone deacetylase 8, ... | | Authors: | Zheng, X, Conti, C, Caravella, J, Zablocki, M.-M, Bair, K, Barczak, N, Han, B, Lancia Jr, D, Liu, C, Martin, M, Ng, P.Y, Rudnitskaya, A, Thomason, J.J, Garcia-Dancey, R, Hardy, C, Lahdenranta, J, Leng, C, Li, P, Pardo, E, Saldahna, A, Tan, T, Toms, A.V, Yao, L, Zhang, C. | | Deposit date: | 2019-03-26 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based Discovery of Novel N-(E)-N-Hydroxy-3-(2-(2-oxoimidazolidin-1-yl)phenyl)acrylamides as Potent and Selective HDAC8 inhibitors
To Be Published
|
|
6O4B
 
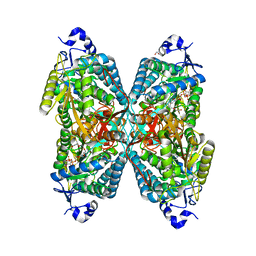 | | Structure of ALDH7A1 mutant W175G complexed with NAD | | Descriptor: | 1,2-ETHANEDIOL, Alpha-aminoadipic semialdehyde dehydrogenase, CHLORIDE ION, ... | | Authors: | Tanner, J.J, Korasick, D.A, Laciak, A.R. | | Deposit date: | 2019-02-28 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and biochemical consequences of pyridoxine-dependent epilepsy mutations that target the aldehyde binding site of aldehyde dehydrogenase ALDH7A1.
Febs J., 287, 2020
|
|
6F3V
 
 | | Backbone structure of bradykinin (BK) peptide bound to human Bradykinin 2 Receptor (B2R) determined by MAS SSNMR | | Descriptor: | Bradykinin (BK) | | Authors: | Mao, J, Lopez, J.J, Shukla, A.K, Kuenze, G, Meiler, J, Schwalbe, H, Michel, H, Glaubitz, C. | | Deposit date: | 2017-11-29 | | Release date: | 2018-01-10 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | The molecular basis of subtype selectivity of human kinin G-protein-coupled receptors.
Nat. Chem. Biol., 14, 2018
|
|
6F3W
 
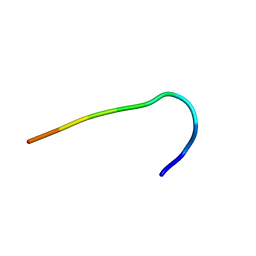 | | Backbone structure of free bradykinin (BK) in DDM/CHS detergent micelle determined by MAS SSNMR | | Descriptor: | Kininogen-1 | | Authors: | Mao, J, Lopez, J.J, Shukla, A.K, Kuenze, G, Meiler, J, Schwalbe, H, Michel, H, Glaubitz, C. | | Deposit date: | 2017-11-29 | | Release date: | 2018-01-10 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | The molecular basis of subtype selectivity of human kinin G-protein-coupled receptors.
Nat. Chem. Biol., 14, 2018
|
|
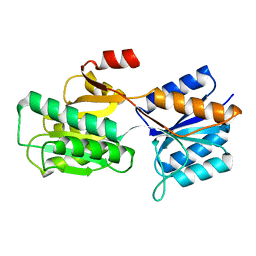
8ABP
 
 | |
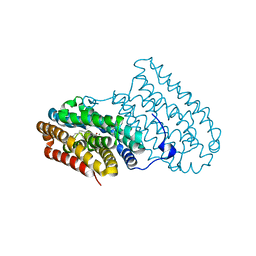
6QJV
 
 | |
6FQ3
 
 | | Crystal structure of Danio rerio Lin41 filamin-NHL domains in complex with lin-29A 5'UTR 13mer RNA | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase TRIM71, RNA (5'-R(*GP*GP*AP*GP*UP*CP*CP*AP*AP*CP*UP*CP*C)-3') | | Authors: | Kumari, P, Aeschimann, F, Gaidatzis, D, Keusch, J.J, Ghosh, P, Neagu, A, Pachulska-Wieczorek, K, Bujnicki, J.M, Gut, H, Grosshans, H, Ciosk, R. | | Deposit date: | 2018-02-13 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Evolutionary plasticity of the NHL domain underlies distinct solutions to RNA recognition.
Nat Commun, 9, 2018
|
|
6QK1
 
 | |
6QRZ
 
 | | Crystal structure of R2-like ligand-binding oxidase from Sulfolobus acidocaldarius solved by 3D micro-crystal electron diffraction | | Descriptor: | FE (III) ION, MANGANESE (III) ION, Ribonucleoside-diphosphate reductase | | Authors: | Xu, H, Lebrette, H, Clabbers, M.T.B, Zhao, J, Griese, J.J, Zou, X, Hogbom, M. | | Deposit date: | 2019-02-20 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | ELECTRON CRYSTALLOGRAPHY (3 Å) | | Cite: | Solving a new R2lox protein structure by microcrystal electron diffraction.
Sci Adv, 5, 2019
|
|
