8BJD
 
 | | Full length structure of LpMIP with bound inhibitor JK095 | | Descriptor: | (1~{S},5~{S},6~{R})-10-[3,5-bis(chloranyl)phenyl]sulfonyl-5-(hydroxymethyl)-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Whittaker, J.J, Guskov, A, Goretzki, B, Hellmich, U.A. | | Deposit date: | 2022-11-04 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Legionella pneumophila macrophage infectivity potentiator protein appendage domains modulate protein dynamics and inhibitor binding.
Int.J.Biol.Macromol., 252, 2023
|
|
8BK4
 
 | | Full length structure of the apo-state LpMIP. | | Descriptor: | (1~{S},5~{S},6~{R})-10-[3,5-bis(chloranyl)phenyl]sulfonyl-5-(hydroxymethyl)-3-[(1~{S})-1-pyridin-2-ylethyl]-3,10-diazabicyclo[4.3.1]decan-2-one, GLYCEROL, Macrophage infectivity potentiator, ... | | Authors: | Whittaker, J.J, Guskov, A, Goretzki, B, Hellmich, U.A. | | Deposit date: | 2022-11-08 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Legionella pneumophila macrophage infectivity potentiator protein appendage domains modulate protein dynamics and inhibitor binding.
Int.J.Biol.Macromol., 252, 2023
|
|
8BK5
 
 | | A structure of the truncated LpMIP with bound inhibitor JK095. | | Descriptor: | (1~{S},5~{S},6~{R})-10-[3,5-bis(chloranyl)phenyl]sulfonyl-5-(hydroxymethyl)-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one, Peptidyl-prolyl cis-trans isomerase, SODIUM ION | | Authors: | Whittaker, J.J, Guskov, A, Goretzki, B, Hellmich, U.A. | | Deposit date: | 2022-11-08 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Legionella pneumophila macrophage infectivity potentiator protein appendage domains modulate protein dynamics and inhibitor binding.
Int.J.Biol.Macromol., 252, 2023
|
|
8BJE
 
 | | A structure of the truncated LpMIP with bound inhibitor JK236. | | Descriptor: | (1~{S},5~{S},6~{R})-10-[3,5-bis(chloranyl)phenyl]sulfonyl-5-(hydroxymethyl)-3-[(1~{S})-1-pyridin-2-ylethyl]-3,10-diazabicyclo[4.3.1]decan-2-one, GLYCEROL, Peptidyl-prolyl cis-trans isomerase | | Authors: | Whittaker, J.J, Guskov, A, Goretzki, B, Hellmich, U.A. | | Deposit date: | 2022-11-04 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Legionella pneumophila macrophage infectivity potentiator protein appendage domains modulate protein dynamics and inhibitor binding.
Int.J.Biol.Macromol., 252, 2023
|
|
8BJC
 
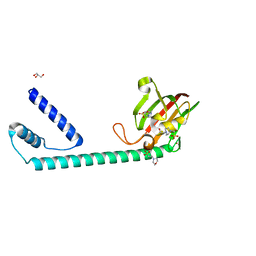 | | Full length structure of the apo-state LpMIP. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Whittaker, J.J, Guskov, A, Goretzki, B, Hellmich, U.A. | | Deposit date: | 2022-11-03 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Legionella pneumophila macrophage infectivity potentiator protein appendage domains modulate protein dynamics and inhibitor binding.
Int.J.Biol.Macromol., 252, 2023
|
|
8FHI
 
 | | Wildtype rabbit TRPV5 in nanodiscs in complex with oleoyl coenzyme A, Open stated | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, ERGOSTEROL, S-{(3R,5R,9R)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-3,5-dioxido-10,14-dioxo-2,4,6-trioxa-11,15-diaza-3lambda~5~,5lambda~5~-diphosphaheptadecan-17-yl} (9Z)-octadec-9-enethioate (non-preferred name), ... | | Authors: | De Jesus-Perez, J.J, Moiseenkova-Bell, V.Y. | | Deposit date: | 2022-12-14 | | Release date: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural basis of the activation of TRPV5 channels by long-chain acyl-Coenzyme-A.
Nat Commun, 14, 2023
|
|
8FFO
 
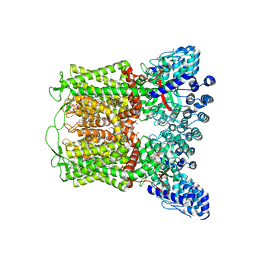 | |
8FHH
 
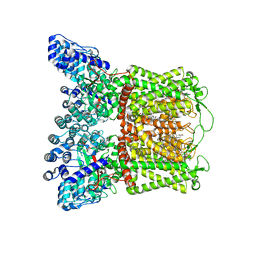 | |
5UZ3
 
 | | Solution Structure of a DNA Dodecamer with 5-methylcytosine at the 9th position and 8-oxoguanine at the 10th position | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*(DMC)P*(8OG)P*CP*G)-3') | | Authors: | Gruber, D.R, Hoppins, J.J, Miears, H.L, Endutkin, A.V, Zharkov, D.O, Smirnov, S.L. | | Deposit date: | 2017-02-24 | | Release date: | 2017-05-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Oxidative damage to epigenetically methylated sites affects DNA stability, dynamics and enzymatic demethylation.
Nucleic Acids Res., 46, 2018
|
|
2BRT
 
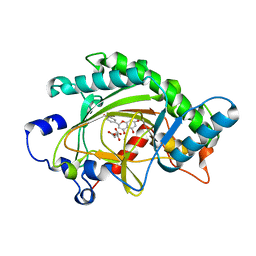 | | ANTHOCYANIDIN SYNTHASE FROM ARABIDOPSIS THALIANA COMPLEXED with naringenin | | Descriptor: | 2-OXOGLUTARIC ACID, FE (II) ION, LEUCOANTHOCYANIDIN DIOXYGENASE, ... | | Authors: | Turnbull, J.J, Clifton, I.J, Welford, R.W.D, Schofield, C.J. | | Deposit date: | 2005-05-11 | | Release date: | 2006-08-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Mechanistic Studies on Anthocyanidin Synthase Catalysed Oxidation of Flavanone Substrates: The Effect of C-2 Stereochemistry on Product Selectivity and Mechanism
Org.Biomol.Chem., 3, 2005
|
|
5UZ2
 
 | | Solution Structure of a DNA Dodecamer with 5-methylcytosine at the 3rd and 9th position and 8-oxoguanine at the 10th position | | Descriptor: | DNA (5'-D(*CP*GP*(DMC)P*GP*AP*AP*TP*TP*(DMC)P*(8OG)P*CP*G)-3') | | Authors: | Gruber, D.R, Hoppins, J.J, Miears, H.L, Endutkin, A.V, Zharkov, D.O, Smirnov, S.L. | | Deposit date: | 2017-02-24 | | Release date: | 2017-03-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Oxidative damage to epigenetically methylated sites affects DNA stability, dynamics and enzymatic demethylation.
Nucleic Acids Res., 46, 2018
|
|
2BWN
 
 | | 5-Aminolevulinate Synthase from Rhodobacter capsulatus | | Descriptor: | 5-AMINOLEVULINATE SYNTHASE, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Astner, I, Schulze, J.O, van den Heuvel, J.J, Jahn, D, Schubert, W.-D, Heinz, D.W. | | Deposit date: | 2005-07-15 | | Release date: | 2005-09-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of 5-Aminolevulinate Synthase, the First Enzyme of Heme Biosynthesis, and its Link to Xlsa in Humans.
Embo J., 24, 2005
|
|
6X9B
 
 | |
8BK6
 
 | | A truncated structure of LpMIP with bound inhibitor JK095. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, Peptidyl-prolyl cis-trans isomerase | | Authors: | Whittaker, J.J, Guskov, A, Hellmich, A.U, Goretzki, B. | | Deposit date: | 2022-11-08 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.263 Å) | | Cite: | Legionella pneumophila macrophage infectivity potentiator protein appendage domains modulate protein dynamics and inhibitor binding.
Int.J.Biol.Macromol., 252, 2023
|
|
2CW0
 
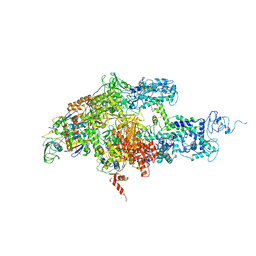 | | Crystal structure of Thermus thermophilus RNA polymerase holoenzyme at 3.3 angstroms resolution | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase beta' chain, ... | | Authors: | Tuske, S, Sarafianos, S.G, Wang, X, Hudson, B, Sineva, E, Mukhopadhyay, J, Birktoft, J.J, Leroy, O, Ismail, S, Clark Jr, A.D, Dharia, C, Napoli, A, Laptenko, O, Lee, J, Borukhov, S, Ebright, R.H, Arnold, E. | | Deposit date: | 2005-06-15 | | Release date: | 2005-09-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Inhibition of bacterial RNA polymerase by streptolydigin: stabilization of a straight-bridge-helix active-center conformation
Cell(Cambridge,Mass.), 122, 2005
|
|
6XP1
 
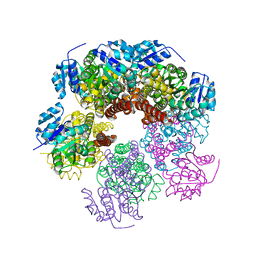 | |
6XE4
 
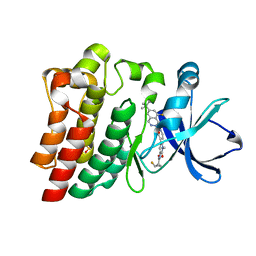 | | BTK Fluorocyclopropyl amide inhibitor, Compound 25 | | Descriptor: | (1S,2S)-N-[2'-(6-tert-butyl-8-fluoro-1-oxophthalazin-2(1H)-yl)-3'-(hydroxymethyl)-1-methyl-6-oxo[1,6-dihydro[3,4'-bipyridine]]-5-yl]-2-fluorocyclopropane-1-carboxamide, SULFATE ION, Tyrosine-protein kinase BTK | | Authors: | Kiefer, J.R, Crawford, J.J, Lee, W, Eigenbrot, C, Yu, C. | | Deposit date: | 2020-06-11 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Stereochemical Differences in Fluorocyclopropyl Amides Enable Tuning of Btk Inhibition and Off-Target Activity.
Acs Med.Chem.Lett., 11, 2020
|
|
6X9C
 
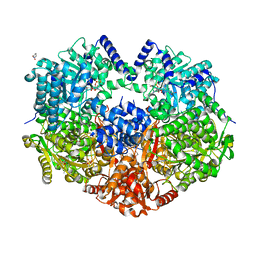 | |
6X99
 
 | |
6XOZ
 
 | |
6X9D
 
 | |
2VBK
 
 | | NATIVE TAILSPIKE PROTEIN OF BACTERIOPHAGE SF6 | | Descriptor: | 1,2-ETHANEDIOL, CARBON DIOXIDE, GLYCEROL, ... | | Authors: | Mueller, J.J, Barbirz, S, Heinle, K, Freiberg, A, Seckler, R, Heinemann, U. | | Deposit date: | 2007-09-14 | | Release date: | 2008-04-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | An intersubunit active site between supercoiled parallel beta helices in the trimeric tailspike endorhamnosidase of Shigella flexneri Phage Sf6.
Structure, 16, 2008
|
|
6XP2
 
 | |
2BBU
 
 | |
2BE9
 
 | | Crystal structure of the CTP-liganded (T-State) aspartate transcarbamoylase from the extremely thermophilic archaeon Sulfolobus acidocaldarius | | Descriptor: | Aspartate carbamoyltransferase, Aspartate carbamoyltransferase regulatory chain, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | De Vos, D, Savvides, S.N, Van Beeumen, J.J. | | Deposit date: | 2005-10-23 | | Release date: | 2006-10-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Sulfolobus acidocaldarius aspartate carbamoyltransferase in complex with its allosteric activator CTP.
Biochem.Biophys.Res.Commun., 372, 2008
|
|
