6ZG8
 
 | | bovine ATP synthase rotor domain state 2 | | Descriptor: | ATP synthase F(0) complex subunit C2, mitochondrial, ATP synthase subunit delta, ... | | Authors: | Spikes, T, Montgomery, M.G, Walker, J.E. | | Deposit date: | 2020-06-18 | | Release date: | 2020-09-09 | | Last modified: | 2020-09-30 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structure of the dimeric ATP synthase from bovine mitochondria.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2XDX
 
 | | Structure of HSP90 with small molecule inhibitor bound | | Descriptor: | 4-CHLORO-6-(2-METHOXYPHENYL)PYRIMIDIN-2-AMINE, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Murray, C.W, Carr, M.G, Callaghan, O, Chessari, G, Congreve, M, Cowan, S, Coyle, J.E, Downham, R, Figueroa, E, Frederickson, M, Graham, B, McMenamin, R, O'Brien, M.A, Patel, S, Phillips, T.R, Williams, G, Woodhead, A.J, Woolford, A.J.A. | | Deposit date: | 2010-05-10 | | Release date: | 2010-09-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Fragment-Based Drug Discovery Applied to Hsp90. Discovery of Two Lead Series with High Ligand Efficiency.
J.Med.Chem., 53, 2010
|
|
6Z1U
 
 | | bovine ATP synthase F1c8-peripheral stalk domain, state 3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase F(0) complex subunit B1, ... | | Authors: | Spikes, T, Montgomery, M.G, Walker, J.E. | | Deposit date: | 2020-05-14 | | Release date: | 2020-09-09 | | Last modified: | 2020-09-30 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structure of the dimeric ATP synthase from bovine mitochondria.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2XJX
 
 | | Structure of HSP90 with small molecule inhibitor bound | | Descriptor: | HEAT SHOCK PROTEIN HSP 90-ALPHA, Onalespib | | Authors: | Murray, C.W, Carr, M.G, Callaghan, O, Chessari, G, Congreve, M, Cowan, S, Coyle, J.E, Downham, R, Figueroa, E, Frederickson, M, Graham, B, McMenamin, R, OBrien, M.A, Patel, S, Phillips, T.R, Williams, G, Woodhead, A.J, Woolford, A.J.A. | | Deposit date: | 2010-07-06 | | Release date: | 2010-08-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Discovery of (2,4-Dihydroxy-5-Isopropylphenyl)-[5-(4-Methylpiperazin-1-Ylmethyl)-1,3-Dihydroisoindol-2-Yl]Methanone (at13387), a Novel Inhibitor of the Molecular Chaperone Hsp90 by Fragment Based Drug Design.
J.Med.Chem., 53, 2010
|
|
4FTB
 
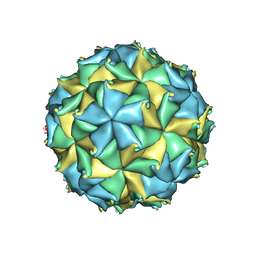 | | Crystal structure of the authentic Flock House virus particle | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Speir, J.A, Chen, Z, Reddy, V.S, Johnson, J.E. | | Deposit date: | 2012-06-27 | | Release date: | 2012-08-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural study of virus assembly intermediates reveals maturation event sequence and a staging position for externalized lytic peptides
to be published, 2012
|
|
1CLD
 
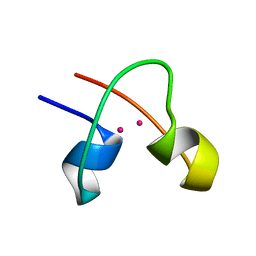 | | DNA-binding protein | | Descriptor: | CADMIUM ION, CD2-LAC9 | | Authors: | Gardner, K.H, Coleman, J.E. | | Deposit date: | 1995-06-06 | | Release date: | 1995-09-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Kluyveromyces lactis LAC9 Cd2 Cys6 DNA-binding domain.
Nat.Struct.Biol., 2, 1995
|
|
5O8A
 
 | | Crystal Structure of rsEGFP2 in the non-fluorescent off-state determined by SFX | | Descriptor: | Green fluorescent protein | | Authors: | Coquelle, N, Sliwa, M, Woodhouse, J, Schiro, G, Adam, V, Aquila, A, Barends, T.R.M, Boutet, S, Byrdin, M, Carbajo, S, De la Mora, E, Doak, R.B, Feliks, M, Fieschi, F, Foucar, L, Guillon, V, Hilpert, M, Hunter, M, Jakobs, S, Koglin, J.E, Kovacsova, G, Lane, T.J, Levy, B, Liang, M, Nass, K, Ridard, J, Robinson, J.S, Roome, C.M, Ruckebusch, C, Seaberg, M, Thepaut, M, Cammarata, M, Demachy, I, Field, M, Shoeman, R.L, Bourgeois, D, Colletier, J.P, Schlichting, I, Weik, M. | | Deposit date: | 2017-06-12 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Chromophore twisting in the excited state of a photoswitchable fluorescent protein captured by time-resolved serial femtosecond crystallography.
Nat Chem, 10, 2018
|
|
5OFV
 
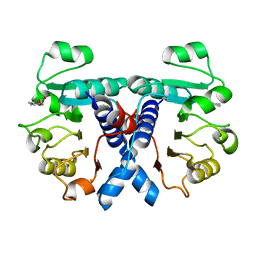 | | Crystal structure of human 3-phosphoglycerate dehydrogenase in complex with 5-fluoro-2-methylbenzoic acid | | Descriptor: | 5-fluoranyl-2-methyl-benzoic acid, D-3-phosphoglycerate dehydrogenase | | Authors: | Unterlass, J.E, Basle, A, Blackburn, T.J, Tucker, J, Cano, C, Noble, M.E.M, Curtin, N.J. | | Deposit date: | 2017-07-11 | | Release date: | 2017-08-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Validating and enabling phosphoglycerate dehydrogenase (PHGDH) as a target for fragment-based drug discovery in PHGDH-amplified breast cancer.
Oncotarget, 9, 2018
|
|
6XH1
 
 | |
5JHA
 
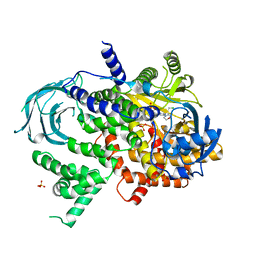 | | Structure of Phosphoinositide 3-kinase gamma (PI3K) bound to the potent inhibitor PIKin2 | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION, [1-{4-[6-amino-4-(trifluoromethyl)pyridin-3-yl]-6-(morpholin-4-yl)pyrimidin-2-yl}-3-(chloromethyl)azetidin-3-yl]methanol | | Authors: | Burke, J.E, Inglis, A.J, Williams, R.L. | | Deposit date: | 2016-04-20 | | Release date: | 2017-03-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Deconvolution of Buparlisib's mechanism of action defines specific PI3K and tubulin inhibitors for therapeutic intervention.
Nat Commun, 8, 2017
|
|
5NZO
 
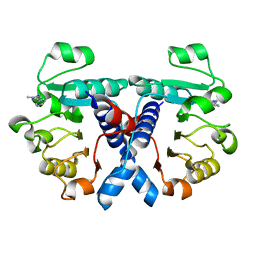 | | Crystal structure of human 3-phosphoglycerate dehydrogenase in complex with 1-methyl-3-phenyl-1H-pyrazol-5-amine | | Descriptor: | 2-methyl-5-phenyl-pyrazol-3-amine, D-3-phosphoglycerate dehydrogenase | | Authors: | Unterlass, J.E, Basle, A, Blackburn, T.J, Tucker, J, Cano, C, Noble, M.E.M, Curtin, N.J. | | Deposit date: | 2017-05-14 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Validating and enabling phosphoglycerate dehydrogenase (PHGDH) as a target for fragment-based drug discovery in PHGDH-amplified breast cancer.
Oncotarget, 9, 2018
|
|
4GOO
 
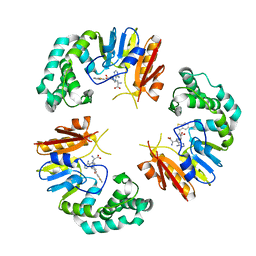 | |
5NXB
 
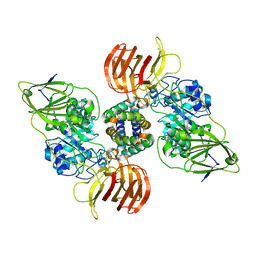 | | Mouse galactocerebrosidase in complex with saposin A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Graham, S.C, Hill, C.H, Deane, J.E. | | Deposit date: | 2017-05-09 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The mechanism of glycosphingolipid degradation revealed by a GALC-SapA complex structure.
Nat Commun, 9, 2018
|
|
4GT2
 
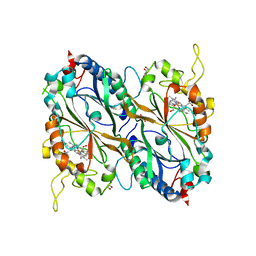 | | Crystal structure of DyP-type peroxidase (SCO3963) from Streptomyces coelicolor | | Descriptor: | ACETATE ION, GLYCEROL, OXYGEN MOLECULE, ... | | Authors: | Lukk, T, Hetta, A.M.A, Jones, A, Solbiati, J, Majumdar, S, Cronan, J.E, Gerlt, J.A, Nair, S.K. | | Deposit date: | 2012-08-28 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | DyP-type peroxidases from Stretptomyces and Thermobifida can modify organosolv lignin.
To be Published
|
|
4GUB
 
 | | Casein Kinase II bound to Inhibitor | | Descriptor: | CHLORIDE ION, Casein kinase II subunit alpha, N-[5-({3-cyano-7-[(1-methyl-1H-imidazol-4-yl)amino]pyrazolo[1,5-a]pyrimidin-5-yl}amino)-2-methylphenyl]acetamide | | Authors: | Larsen, N.A, Dowling, J.E. | | Deposit date: | 2012-08-29 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Potent and selective inhibitors of CK2 kinase.
To be Published
|
|
4HAQ
 
 | | Crystal Structure of a GH7 family cellobiohydrolase from Limnoria quadripunctata in complex with cellobiose and cellotriose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, GH7 family protein, ... | | Authors: | Martin, R.N.A, McGeehan, J.E, Streeter, S.D, Cragg, S.M, Guille, M.J, Schnorr, K.M, Kern, M, Bruce, N.C, McQueen-Mason, S.J. | | Deposit date: | 2012-09-27 | | Release date: | 2013-06-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterization of a unique marine animal family 7 cellobiohydrolase suggests a mechanism of cellulase salt tolerance
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3MXF
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor JQ1 | | Descriptor: | (6S)-6-(2-tert-butoxy-2-oxoethyl)-4-(4-chlorophenyl)-2,3,9-trimethyl-6,7-dihydrothieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-10-ium, 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ... | | Authors: | Filippakopoulos, P, Picaud, S, Qi, J, Keates, T, Felletar, I, Fedorov, O, Muniz, J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Bradner, J.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-05-07 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Selective inhibition of BET bromodomains.
Nature, 468, 2010
|
|
2ZFC
 
 | | X-ray crystal structure of an engineered N-terminal HIV-1 GP41 trimer with enhanced stability and potency | | Descriptor: | HIV-1 GP41 | | Authors: | Dwyer, J.J, Wilson, K.L, Martin, K, Seedorff, J.E, Hasan, A, Kim, H. | | Deposit date: | 2007-12-29 | | Release date: | 2008-04-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Design of an engineered N-terminal HIV-1 gp41 trimer with enhanced stability and potency
Protein Sci., 17, 2008
|
|
5NZQ
 
 | | Crystal structure of human 3-phosphoglycerate dehydrogenase in complex with 3-(1,3-oxazol-5-yl)aniline. | | Descriptor: | 3-(1,3-oxazol-5-yl)aniline, D-3-phosphoglycerate dehydrogenase | | Authors: | Unterlass, J.E, Basle, A, Blackburn, T.J, Tucker, J, Cano, C, Noble, M.E.M, Curtin, N.J. | | Deposit date: | 2017-05-14 | | Release date: | 2017-06-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Validating and enabling phosphoglycerate dehydrogenase (PHGDH) as a target for fragment-based drug discovery in PHGDH-amplified breast cancer.
Oncotarget, 9, 2018
|
|
6VY6
 
 | |
4GRH
 
 | | Crystal structure of pabB of Stenotrophomonas maltophilia | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, MAGNESIUM ION, POLYETHYLENE GLYCOL (N=34), ... | | Authors: | Bera, A, Atanasova, V, Ladner, J.E, Parsons, J.F. | | Deposit date: | 2012-08-24 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of Aminodeoxychorismate Synthase from Stenotrophomonas maltophilia.
Biochemistry, 51, 2012
|
|
5IC4
 
 | | Crystal structure of caspase-3 DEVE peptide complex | | Descriptor: | Caspase-3 subunit p12, Caspase-3 subunit p17, DEVE peptide | | Authors: | Seaman, J.E, Julien, O, Lee, P.S, Rettenmaier, T.J, Thomsen, N.D, Wells, J.A. | | Deposit date: | 2016-02-22 | | Release date: | 2016-07-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Cacidases: caspases can cleave after aspartate, glutamate and phosphoserine residues.
Cell Death Differ., 23, 2016
|
|
6ZIU
 
 | | bovine ATP synthase stator domain, state 3 | | Descriptor: | ATP synthase F(0) complex subunit B1, mitochondrial, ATP synthase protein 8, ... | | Authors: | Spikes, T.E, Montgomery, M.G, Walker, J.E. | | Deposit date: | 2020-06-26 | | Release date: | 2020-09-09 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (6.02 Å) | | Cite: | Structure of the dimeric ATP synthase from bovine mitochondria.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5IK2
 
 | | Caldalaklibacillus thermarum F1-ATPase (epsilon mutant) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP synthase epsilon chain, ATP synthase gamma chain, ... | | Authors: | Ferguson, S.A, Cook, G.M, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2016-03-03 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Regulation of the thermoalkaliphilic F1-ATPase from Caldalkalibacillus thermarum.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6VY5
 
 | |
