6SMB
 
 | | Human jak1 kinase domain in complex with inhibitor | | 分子名称: | Tyrosine-protein kinase JAK1, ~{N}-[3-[2-[(3-methoxy-1-methyl-pyrazol-4-yl)amino]-5-methyl-pyrimidin-4-yl]-1~{H}-indol-7-yl]-2-methyl-pyridine-3-carboxamide | | 著者 | Read, J.A, Steuber, H. | | 登録日 | 2019-08-21 | | 公開日 | 2020-04-29 | | 最終更新日 | 2020-05-27 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Discovery of (2R)-N-[3-[2-[(3-Methoxy-1-methyl-pyrazol-4-yl)amino]pyrimidin-4-yl]-1H-indol-7-yl]-2-(4-methylpiperazin-1-yl)propenamide (AZD4205) as a Potent and Selective Janus Kinase 1 Inhibitor.
J.Med.Chem., 63, 2020
|
|
6TWW
 
 | | Variant W229D/F290W-19 of the last common ancestor of Gram-negative bacteria beta-lactamase class A (GNCA4) | | 分子名称: | ACETATE ION, Beta-Lactamase (GNCA4), FORMIC ACID, ... | | 著者 | Gavira, J.A, Risso, V, Sanchez-Ruiz, J.M, Romero-Rivera, A, Kamerlin, S.C.L. | | 登録日 | 2020-01-13 | | 公開日 | 2020-06-03 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | Enhancing ade novoenzyme activity by computationally-focused ultra-low-throughput screening.
Chem Sci, 11, 2020
|
|
2HDL
 
 | | Solution structure of Brak/CXCL14 | | 分子名称: | Small inducible cytokine B14 | | 著者 | Peterson, F.C, Thorpe, J.A, Harder, A.G, Volkman, B.F, Schwarze, S.R. | | 登録日 | 2006-06-20 | | 公開日 | 2006-10-24 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Determinants Involved in the Regulation of CXCL14/BRAK Expression by the 26 S Proteasome.
J.Mol.Biol., 363, 2006
|
|
6U8K
 
 | | Crystal structure of hepatitis C virus IRES junction IIIabc in complex with Fab HCV3 | | 分子名称: | Heavy chain of Fab HCV3, JIIIabc RNA (68-MER), Light chain of Fab HCV3 | | 著者 | Koirala, D, Lewicka, A, Koldobskaya, Y, Huang, H, Piccirilli, J.A. | | 登録日 | 2019-09-05 | | 公開日 | 2019-12-04 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Synthetic Antibody Binding to a Preorganized RNA Domain of Hepatitis C Virus Internal Ribosome Entry Site Inhibits Translation.
Acs Chem.Biol., 15, 2020
|
|
2HI9
 
 | |
6U63
 
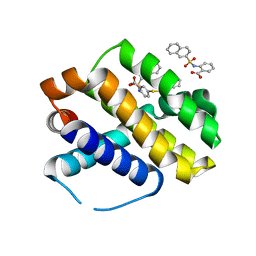 | | Mcl-1 bound to compound 17 | | 分子名称: | 2-{[(naphthalen-2-yl)sulfonyl]amino}-5-[(2-phenylethyl)sulfanyl]benzoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | 著者 | Stuckey, J.A. | | 登録日 | 2019-08-29 | | 公開日 | 2020-03-04 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Discovery and Characterization of 2,5-Substituted Benzoic Acid Dual Inhibitors of the Anti-apoptotic Mcl-1 and Bfl-1 Proteins.
J.Med.Chem., 63, 2020
|
|
6TXD
 
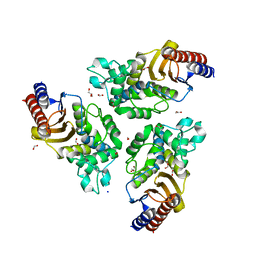 | | Variant W229D/F290W-12 of the last common ancestor of Gram-negative bacteria beta-lactamase class A (GNCA4) | | 分子名称: | ACETATE ION, Beta lactamase (GNCA4-12), FORMIC ACID, ... | | 著者 | Gavira, J.A, Risso, V, Sanchez-Ruiz, J.M, Romero-Rivera, A, Kamerlin, S.C.L. | | 登録日 | 2020-01-14 | | 公開日 | 2020-06-03 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Enhancing ade novoenzyme activity by computationally-focused ultra-low-throughput screening.
Chem Sci, 11, 2020
|
|
6TQ6
 
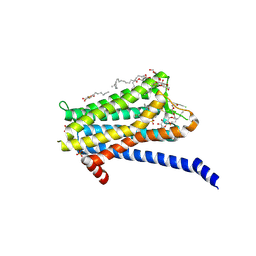 | | Crystal structure of the Orexin-1 receptor in complex with Compound 14 | | 分子名称: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, 2-(5-methylsulfonylpyridin-3-yl)-1,1-bis(oxidanylidene)-4-[[2,4,6-tris(fluoranyl)phenyl]methyl]pyrido[2,3-e][1,2,4]thiadiazin-3-one, Orexin receptor type 1, ... | | 著者 | Rappas, M, Ali, A, Bennett, K.A, Brown, J.D, Bucknell, S.J, Congreve, M, Cooke, R.M, Cseke, G, de Graaf, C, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Mason, J.S, Mould, R, Patel, J.C, Tehan, B.G, Weir, M, Christopher, J.A. | | 登録日 | 2019-12-16 | | 公開日 | 2020-01-01 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.546 Å) | | 主引用文献 | Comparison of Orexin 1 and Orexin 2 Ligand Binding Modes Using X-ray Crystallography and Computational Analysis.
J.Med.Chem., 63, 2020
|
|
6U9K
 
 | | MLL1 SET N3861I/Q3867L bound to inhibitor 18 (TC-5153) | | 分子名称: | 5'-([(3S)-3-amino-3-carboxypropyl]{[1-(3,3-diphenylpropyl)azetidin-3-yl]methyl}amino)-5'-deoxyadenosine, GLYCEROL, Histone-lysine N-methyltransferase, ... | | 著者 | Petrunak, E.M, Stuckey, J.A. | | 登録日 | 2019-09-09 | | 公開日 | 2020-07-01 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Discovery of Potent Small-Molecule Inhibitors of MLL Methyltransferase.
Acs Med.Chem.Lett., 11, 2020
|
|
6U6F
 
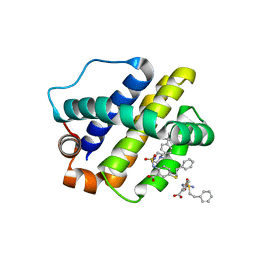 | | The crystal structure of anti-apoptotic Mcl-1 protein in complex with 2, 5-substituted benzoic acid inhibitor 21 | | 分子名称: | 2-[({4-[(4-tert-butylphenyl)methyl]piperazin-1-yl}sulfonyl)amino]-5-[(2-phenylethyl)sulfanyl]benzoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | 著者 | Yang, Y, Stuckey, J.A, Nikolovska-Coleska, Z. | | 登録日 | 2019-08-29 | | 公開日 | 2020-03-04 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Discovery and Characterization of 2,5-Substituted Benzoic Acid Dual Inhibitors of the Anti-apoptotic Mcl-1 and Bfl-1 Proteins.
J.Med.Chem., 63, 2020
|
|
2K57
 
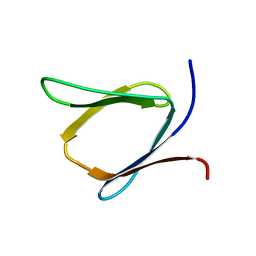 | | Solution NMR Structure of Putative Lipoprotein from Pseudomonas syringae Gene Locus PSPTO2350. Northeast Structural Genomics Target PsR76A. | | 分子名称: | Putative Lipoprotein | | 著者 | Hang, D, Aramini, J.A, Rossi, P, Wang, D, Jiang, M, Maglaqui, M, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2008-06-25 | | 公開日 | 2008-09-30 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution NMR Structure of Putative Lipoprotein from
Pseudomonas syringae Gene Locus PSPTO2350. Northeast Structural Genomics Target PsR76A.
To be Published
|
|
2HZ2
 
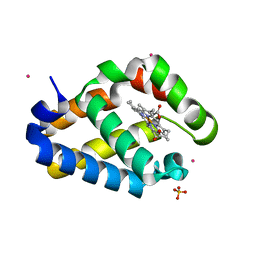 | |
2HZ3
 
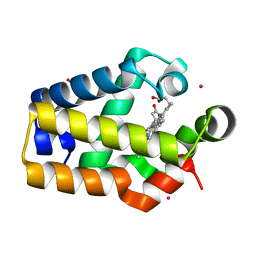 | |
2I5Q
 
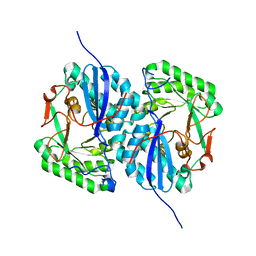 | | Crystal structure of Apo L-rhamnonate dehydratase from Escherichia Coli | | 分子名称: | L-rhamnonate dehydratase | | 著者 | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2006-08-25 | | 公開日 | 2006-09-05 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Evolution of enzymatic activities in the enolase superfamily: L-rhamnonate dehydratase.
Biochemistry, 47, 2008
|
|
2HXU
 
 | | Crystal structure of K220A mutant of L-Fuconate Dehydratase from Xanthomonas campestris liganded with Mg++ and L-fuconate | | 分子名称: | 6-deoxy-L-galactonic acid, L-fuconate dehydratase, MAGNESIUM ION, ... | | 著者 | Fedorov, A.A, Fedorov, E.V, Yew, W.S, Rakus, J.F, Gerlt, J.A, Almo, S.C. | | 登録日 | 2006-08-03 | | 公開日 | 2006-12-19 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Evolution of Enzymatic Activities in the Enolase Superfamily: l-Fuconate Dehydratase from Xanthomonas campestris.
Biochemistry, 45, 2006
|
|
6T92
 
 | | NAD+-dependent fungal formate dehydrogenase from Chaetomium thermophilum: A complex of N120C mutant protein with the reduced form of the cofactor NADH and the substrate formate at a secondary site. | | 分子名称: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Isupov, M.N, Yelmazer, B, De Rose, S.A, Littlechild, J.A. | | 登録日 | 2019-10-25 | | 公開日 | 2020-11-18 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.12 Å) | | 主引用文献 | Structural insights into the NAD + -dependent formate dehydrogenase mechanism revealed from the NADH complex and the formate NAD + ternary complex of the Chaetomium thermophilum enzyme.
J.Struct.Biol., 212, 2020
|
|
6T8Z
 
 | | NAD+-dependent fungal formate dehydrogenase from Chaetomium thermophilum: A ternary complex with the oxidised form of the cofactor NAD+ and the substrate formate both at a primary and secondary sites. | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | 著者 | Isupov, M.N, Yelmazer, B, De Rose, S.A, Littlechild, J.A. | | 登録日 | 2019-10-25 | | 公開日 | 2020-11-18 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.21 Å) | | 主引用文献 | Structural insights into the NAD + -dependent formate dehydrogenase mechanism revealed from the NADH complex and the formate NAD + ternary complex of the Chaetomium thermophilum enzyme.
J.Struct.Biol., 212, 2020
|
|
2HGD
 
 | |
2HGY
 
 | |
2HG6
 
 | | Solution NMR Structure of Protein PA1123 from Pseudomonas aeruginosa. Northeast Structural Genomics Consortium Target PaT4; Ontario Centre for Structural Proteomics Target PA1123. | | 分子名称: | Hypothetical protein | | 著者 | Lemak, A, Srisailam, S, Yee, A, Lukin, J.A, Orekhov, V.Y, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2006-06-26 | | 公開日 | 2006-07-25 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure of a hypothetical protein from Pseudomonas aeruginosa (Northeast Structural Genomics Consortium Target: PaT4; Ontario Centre for Structural Proteomics Target: PA1123)
To be Published
|
|
6S2X
 
 | |
2HI2
 
 | | Crystal structure of native Neisseria gonorrhoeae Type IV pilin at 2.3 Angstroms Resolution | | 分子名称: | Fimbrial protein, HEPTANE-1,2,3-TRIOL, PHOSPHORIC ACID MONO-(2-AMINO-ETHYL) ESTER, ... | | 著者 | Craig, L, Arvai, A.S, Tainer, J.A. | | 登録日 | 2006-06-28 | | 公開日 | 2006-09-12 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Type IV Pilus Structure by Cryo-Electron Microscopy and Crystallography: Implications for Pilus Assembly and Functions.
Mol.Cell, 23, 2006
|
|
6S7P
 
 | | Nucleotide bound ABCB4 | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, CHOLESTEROL, MAGNESIUM ION, ... | | 著者 | Olsen, J.A, Alam, A, Kowal, J, Stieger, B, Locher, K.P. | | 登録日 | 2019-07-05 | | 公開日 | 2019-12-25 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structure of the human lipid exporter ABCB4 in a lipid environment.
Nat.Struct.Mol.Biol., 27, 2020
|
|
2HIU
 
 | | NMR STRUCTURE OF HUMAN INSULIN IN 20% ACETIC ACID, ZINC-FREE, 10 STRUCTURES | | 分子名称: | INSULIN | | 著者 | Hua, Q.X, Gozani, S.N, Chance, R.E, Hoffmann, J.A, Frank, B.H, Weiss, M.A. | | 登録日 | 1996-10-08 | | 公開日 | 1997-04-01 | | 最終更新日 | 2017-11-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of a protein in a kinetic trap.
Nat.Struct.Biol., 2, 1995
|
|
2HRS
 
 | | Crystal structure of L42H V224H design intermediate for GFP metal ion reporter | | 分子名称: | 1,2-ETHANEDIOL, Green fluorescent protein, MAGNESIUM ION | | 著者 | Barondeau, D.P, Tubbs, J.L, Tainer, J.A, Getzoff, E.D. | | 登録日 | 2006-07-20 | | 公開日 | 2008-04-08 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Iterative Structure-Based Design of a Green Fluorescent Protein Metal Ion Reporter
To be Published
|
|
