1YPT
 
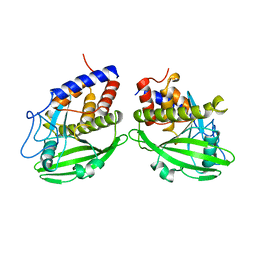 | | CRYSTAL STRUCTURE OF YERSINIA PROTEIN TYROSINE PHOSPHATASE AT 2.5 ANGSTROMS AND THE COMPLEX WITH TUNGSTATE | | Descriptor: | PROTEIN-TYROSINE PHOSPHATASE YERSINIA (CATALYTIC DOMAIN) | | Authors: | Stuckey, J.A, Schubert, H.L, Fauman, E.B, Zhang, Z.-Y, Dixon, J.E, Saper, M.A. | | Deposit date: | 1994-09-16 | | Release date: | 1994-12-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Yersinia protein tyrosine phosphatase at 2.5 A and the complex with tungstate.
Nature, 370, 1994
|
|
6R5I
 
 | |
1EYV
 
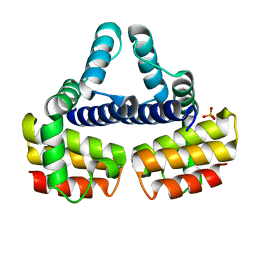 | | THE CRYSTAL STRUCTURE OF NUSB FROM MYCOBACTERIUM TUBERCULOSIS | | Descriptor: | N-UTILIZING SUBSTANCE PROTEIN B HOMOLOG, PHOSPHATE ION | | Authors: | Gopal, B, Haire, L.F, Cox, R.A, Colston, M.J, Major, S, Brannigan, J.A, Smerdon, S.J, Dodson, G.G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2000-05-09 | | Release date: | 2000-05-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of NusB from Mycobacterium tuberculosis.
Nat.Struct.Biol., 7, 2000
|
|
4RZD
 
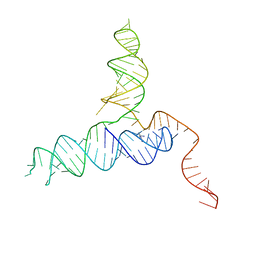 | | Crystal Structure of a PreQ1 Riboswitch | | Descriptor: | 7-DEAZA-7-AMINOMETHYL-GUANINE, PreQ1-III Riboswitch (Class 3) | | Authors: | Wedekind, J.E, Liberman, J.A, Salim, M. | | Deposit date: | 2014-12-20 | | Release date: | 2015-07-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural analysis of a class III preQ1 riboswitch reveals an aptamer distant from a ribosome-binding site regulated by fast dynamics.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6RA7
 
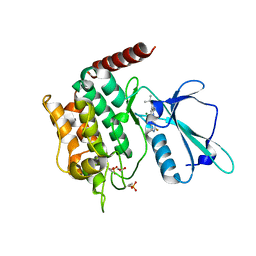 | | Human tnik in complex with compound 9 | | Descriptor: | 1,2-ETHANEDIOL, 2-[8-azanyl-2-(2-fluoranylpyridin-4-yl)-1,7-naphthyridin-5-yl]propan-2-ol, MAGNESIUM ION, ... | | Authors: | Read, J.A. | | Deposit date: | 2019-04-05 | | Release date: | 2019-11-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Tool inhibitors and assays to interrogate the biology of the TRAF2 and NCK interacting kinase
To Be Published
|
|
6RA5
 
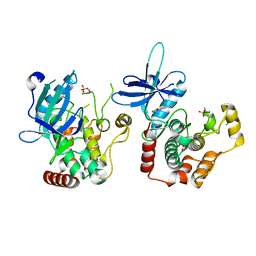 | | Human tnik in complex with compound 9 | | Descriptor: | 5-bromanyl-2-(2-fluoranylpyridin-4-yl)-1,7-naphthyridin-8-amine, MAGNESIUM ION, TRAF2 and NCK-interacting protein kinase | | Authors: | Read, J.A. | | Deposit date: | 2019-04-05 | | Release date: | 2020-05-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Tool inhibitors and assays to interrogate the biology of the TRAF2 and NCK interacting kinase
To Be Published
|
|
5JH1
 
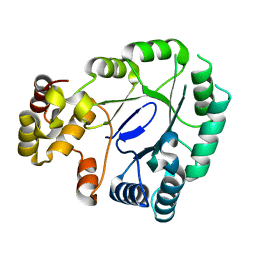 | | Crystal structure of the apo form of AKR4C7 from maize | | Descriptor: | Aldose reductase, AKR4C7 | | Authors: | Giuseppe, P.O, Santos, M.L, Sousa, S.M, Koch, K.E, Yunes, J.A, Aparicio, R, Murakami, M.T. | | Deposit date: | 2016-04-20 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A comparative structural analysis reveals distinctive features of co-factor binding and substrate specificity in plant aldo-keto reductases.
Biochem.Biophys.Res.Commun., 474, 2016
|
|
4RK9
 
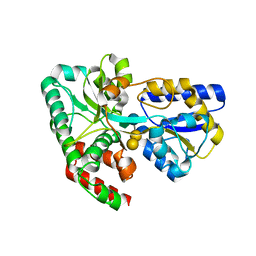 | | CRYSTAL STRUCTURE OF SUGAR TRANSPORTER BL01359 FROM Bacillus licheniformis, TARGET EFI-510856, IN COMPLEX WITH STACHYOSE | | Descriptor: | Carbohydrate ABC transporter substrate-binding protein MsmE, alpha-D-galactopyranose-(1-6)-alpha-D-galactopyranose-(1-6)-[beta-D-fructofuranose-(2-1)]alpha-D-glucopyranose | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Siedel, R.D, Hillerich, B, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-10-12 | | Release date: | 2014-11-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | CRYSTAL STRUCTURE OF SUGAR TRANSPORTER BL01359 FROM Bacillus licheniformis, TARGET EFI-510856
To be Published
|
|
1ET6
 
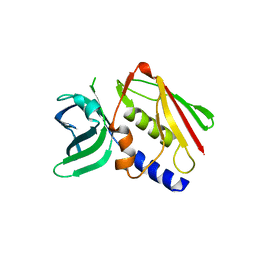 | | CRYSTAL STRUCTURE OF THE SUPERANTIGEN SMEZ-2 FROM STREPTOCOCCUS PYOGENES | | Descriptor: | SUPERANTIGEN SMEZ-2 | | Authors: | Arcus, V.L, Proft, T, Sigrell, J.A, Baker, H.M, Fraser, J.D, Baker, E.N. | | Deposit date: | 2000-04-12 | | Release date: | 2000-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conservation and variation in superantigen structure and activity highlighted by the three-dimensional structures of two new superantigens from Streptococcus pyogenes.
J.Mol.Biol., 299, 2000
|
|
4RY8
 
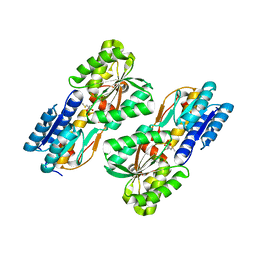 | | Crystal structure of 5-methylthioribose transporter solute binding protein TLET_1677 from Thermotoga lettingae TMO TARGET EFI-511109 in complex with 5-methylthioribose | | Descriptor: | 5-S-methyl-5-thio-alpha-D-ribofuranose, Periplasmic binding protein | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al obaidi, N, Morisco, L.L, Wasserman, S.R, Chamala, S, Attonito, J.D, Scott glenn, A, Chowdhury, S, Lafleur, J, Hillerich, B, Siede, R.D, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-12-13 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of 5-methylthioribose binding protein TLET_1677 from Thermotoga lettingae TARGET EFI-511109
To be Published
|
|
6T95
 
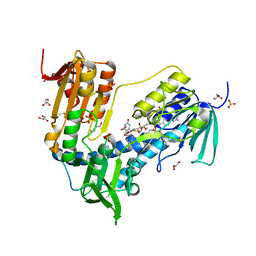 | | Trypanothione Reductase from Leismania infantum in complex with 4a | | Descriptor: | 1-[2-[5-[4-(4-azanylbutyl)-3-methyl-1,2,3-triazol-3-ium-1-yl]-2-[4-(2-phenylethyl)-1,3-thiazol-2-yl]phenoxy]ethyl]imidazolidin-2-one, DIMETHYL SULFOXIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Carriles, A.A, Hermoso, J.A. | | Deposit date: | 2019-10-25 | | Release date: | 2020-11-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of 1,2,3-triazolium salt-based inhibitors of Leishmania infantum trypanothione disulfide reductase with enhanced antileishmanial potency in cellulo and increased selectivity.
Eur.J.Med.Chem., 244, 2022
|
|
1EAL
 
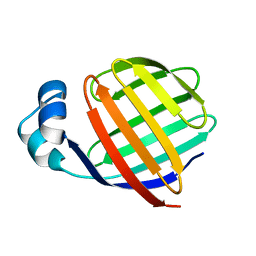 | | NMR STUDY OF ILEAL LIPID BINDING PROTEIN | | Descriptor: | ILEAL LIPID BINDING PROTEIN | | Authors: | Luecke, C, Zhang, F, Rueterjans, H, Hamilton, J.A, Sacchettini, J.C. | | Deposit date: | 1996-08-28 | | Release date: | 1997-01-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Flexibility is a likely determinant of binding specificity in the case of ileal lipid binding protein.
Structure, 4, 1996
|
|
6TP6
 
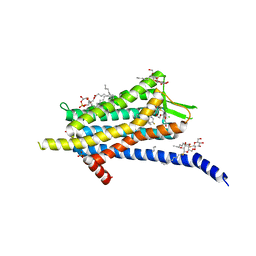 | | Crystal structure of the Orexin-1 receptor in complex with filorexant | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, CHLORIDE ION, Orexin receptor type 1, ... | | Authors: | Rappas, M, Ali, A, Bennett, K.A, Brown, J.D, Bucknell, S.J, Congreve, M, Cooke, R.M, Cseke, G, de Graaf, C, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Mason, J.S, Mould, R, Patel, J.C, Tehan, B.G, Weir, M, Christopher, J.A. | | Deposit date: | 2019-12-12 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.338 Å) | | Cite: | Comparison of Orexin 1 and Orexin 2 Ligand Binding Modes Using X-ray Crystallography and Computational Analysis.
J.Med.Chem., 63, 2020
|
|
6TQ4
 
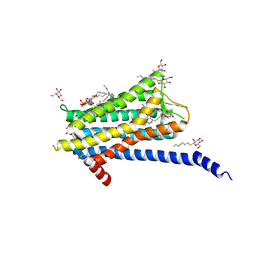 | | Crystal structure of the Orexin-1 receptor in complex with Compound 16 | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, 2-[1-(phenylsulfonyl)-1,8-diazaspiro[4.5]decan-8-yl]-1,3-benzoxazole, Orexin receptor type 1, ... | | Authors: | Rappas, M, Ali, A, Bennett, K.A, Brown, J.D, Bucknell, S.J, Congreve, M, Cooke, R.M, Cseke, G, de Graaf, C, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Mason, J.S, Mould, R, Patel, J.C, Tehan, B.G, Weir, M, Christopher, J.A. | | Deposit date: | 2019-12-16 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Comparison of Orexin 1 and Orexin 2 Ligand Binding Modes Using X-ray Crystallography and Computational Analysis.
J.Med.Chem., 63, 2020
|
|
6T97
 
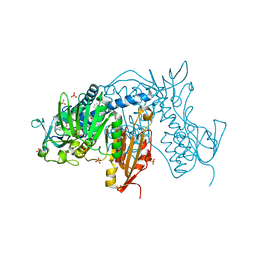 | | Trypanothione Reductase from Leismania infantum in complex with TRL190 | | Descriptor: | 4-[1-[4-[4-(2-phenylethyl)-1,3-thiazol-2-yl]-3-(2-piperidin-4-ylethoxy)phenyl]-1,2,3-triazol-4-yl]butan-1-amine, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Carriles, A, Hermoso, J.A. | | Deposit date: | 2019-10-26 | | Release date: | 2020-11-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Scaffold hopping identifies new triazole-and triazolium-based inhibitors of Leishmania infantum Trypanothione Reductase with potent and selective antileishmanial activity
To Be Published
|
|
4RS3
 
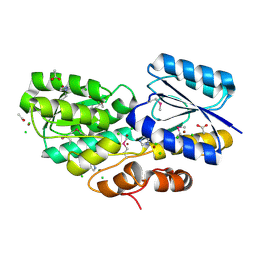 | | Crystal structure of carbohydrate transporter A0QYB3 from Mycobacterium smegmatis str. MC2 155, target EFI-510969, in complex with xylitol | | Descriptor: | ABC transporter, carbohydrate uptake transporter-2 (CUT2) family, periplasmic sugar-binding protein, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Morisco, L.L, Wasserman, S.R, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Hillerich, B, Siedel, R.D, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-11-06 | | Release date: | 2014-11-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A General Strategy for the Discovery of Metabolic Pathways: d-Threitol, l-Threitol, and Erythritol Utilization in Mycobacterium smegmatis.
J.Am.Chem.Soc., 137, 2015
|
|
4RSM
 
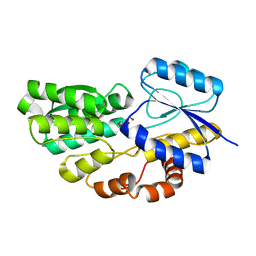 | | Crystal structure of carbohydrate transporter msmeg_3599 from mycobacterium smegmatis str. mc2 155, target efi-510970, in complex with d-threitol | | Descriptor: | D-Threitol, Periplasmic binding protein/LacI transcriptional regulator | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Morisco, L.L, Wasserman, S.R, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Hillerich, B, Siedel, R.D, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-11-08 | | Release date: | 2014-12-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A General Strategy for the Discovery of Metabolic Pathways: d-Threitol, l-Threitol, and Erythritol Utilization in Mycobacterium smegmatis.
J.Am.Chem.Soc., 137, 2015
|
|
5JFT
 
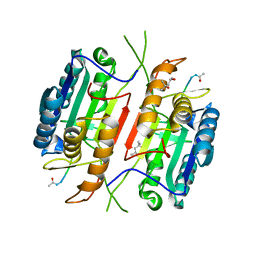 | | Zebra Fish Caspase-3 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ACE-ASP-GLU-VAL-ASK, ... | | Authors: | Tucker, M.B, MacKenzie, S.H, Maciag, J.J, Dirscherl, H, Swartz, P.D, Yoder, J.A, Hamilton, P.T, Clark, A.C. | | Deposit date: | 2016-04-19 | | Release date: | 2016-10-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Phage display and structural studies reveal plasticity in substrate specificity of caspase-3a from zebrafish.
Protein Sci., 25, 2016
|
|
6R5N
 
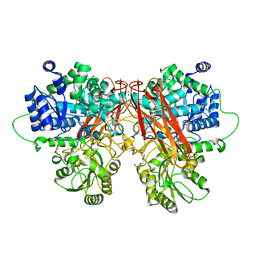 | |
5J8K
 
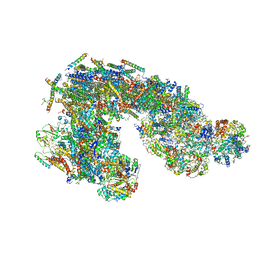 | | Architecture of supercomplex I-III2 | | Descriptor: | COMPLEX I 13KDA/NDUFS6, COMPLEX I 15KDA/NDUFS5, COMPLEX I 18KDA/NDUFS6, ... | | Authors: | Letts, J.A, Fiedorczuk, K, Sazanov, L.A. | | Deposit date: | 2016-04-08 | | Release date: | 2016-09-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | The architecture of respiratory supercomplexes.
Nature, 537, 2016
|
|
2A7Q
 
 | | Crystal structure of human dCK complexed with clofarabine and ADP | | Descriptor: | 2-CHLORO-9-(2-DEOXY-2-FLUORO-B -D-ARABINOFURANOSYL)-9H-PURIN-6-AMINE, ADENOSINE-5'-DIPHOSPHATE, Deoxycytidine kinase, ... | | Authors: | Zhang, Y, Secrist III, J.A, Ealick, S.E. | | Deposit date: | 2005-07-05 | | Release date: | 2006-01-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The structure of human deoxycytidine kinase in complex with clofarabine reveals key interactions for prodrug activation.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
5JGW
 
 | | Crystal structure of maize AKR4C13 in complex with NADP and acetate | | Descriptor: | ACETATE ION, Aldose reductase, AKR4C13, ... | | Authors: | Santos, M.L, Giuseppe, P.O, Kiyota, E, Sousa, S.M, Schmelz, E.A, Yunes, J.A, Koch, K.E, Murakami, M.T, Aparicio, R. | | Deposit date: | 2016-04-20 | | Release date: | 2017-05-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of maize AKR4C13 in complex with NADP and acetate
To Be Published
|
|
1F4D
 
 | | CRYSTAL STRUCTURE OF E. COLI THYMIDYLATE SYNTHASE C146S, L143C COVALENTLY MODIFIED AT C143 WITH N-[TOSYL-D-PROLINYL]AMINO-ETHANETHIOL | | Descriptor: | GLYCEROL, N-[TOSYL-D-PROLINYL]AMINO-ETHANETHIOL, SULFATE ION, ... | | Authors: | Erlanson, D.A, Braisted, A.C, Raphael, D.R, Randal, M, Stroud, R.M, Gordon, E, Wells, J.A. | | Deposit date: | 2000-06-07 | | Release date: | 2000-06-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Site-directed ligand discovery.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1EZX
 
 | |
4TY0
 
 | | Crystal structure of Vibrio cholerae DncV cyclic AMP-GMP synthase in complex with linear intermediate 5' pppA(3',5')pG | | Descriptor: | ACETATE ION, Cyclic AMP-GMP synthase, MAGNESIUM ION, ... | | Authors: | Kranzusch, P.J, Lee, A.S.Y, Wilson, S.C, Solovykh, M.S, Vance, R.E, Berger, J.M, Doudna, J.A. | | Deposit date: | 2014-07-07 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Guided Reprogramming of Human cGAS Dinucleotide Linkage Specificity.
Cell, 158, 2014
|
|
