8AIW
 
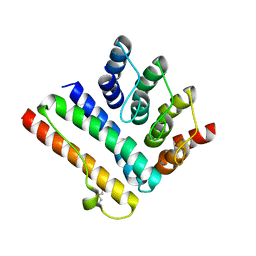 | | Structure of the K5/CagI complex | | Descriptor: | Cag pathogenicity island protein (Cag19), Designed Ankyrin Repeat Protein K5 | | Authors: | Blanc, M, Guerin, J, Terradot, L. | | Deposit date: | 2022-07-27 | | Release date: | 2023-05-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Designed Ankyrin Repeat Proteins provide insights into the structure and function of CagI and are potent inhibitors of CagA translocation by the Helicobacter pylori type IV secretion system.
Plos Pathog., 19, 2023
|
|
8IQX
 
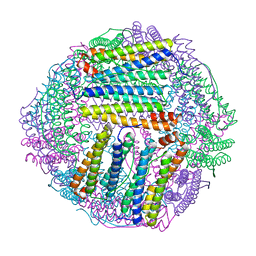 | | ferritin mutant-P156H | | Descriptor: | Ferritin | | Authors: | Zhao, G, Zhang, C, Zang, J, Zhang, T. | | Deposit date: | 2023-03-17 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Preparation and Unique Three-Dimensional Self-Assembly Property of Starfish Ferritin.
Foods, 12, 2023
|
|
1FXQ
 
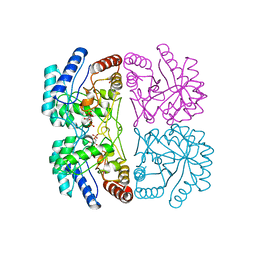 | | AQUIFEX AEOLICUS KDO8P SYNTHASE IN COMPLEX WITH PEP AND A5P | | Descriptor: | 2-DEHYDRO-3-DEOXYPHOSPHOOCTONATE ALDOLASE, ARABINOSE-5-PHOSPHATE, PHOSPHOENOLPYRUVATE | | Authors: | Duewel, H.S, Radaev, S, Wang, J, Woodard, R.W, Gatti, D.L. | | Deposit date: | 2000-09-26 | | Release date: | 2001-04-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Substrate and metal complexes of 3-deoxy-D-manno-octulosonate-8-phosphate synthase from Aquifex aeolicus at 1.9-A resolution. Implications for the condensation mechanism.
J.Biol.Chem., 276, 2001
|
|
6BN6
 
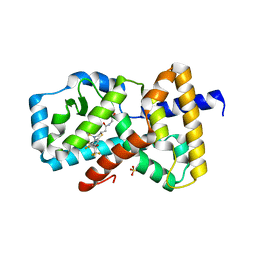 | | IDENTIFICATION OF BICYCLIC HEXAFLUOROISOPROPYL ALCOHOL SULFONAMIDES AS RORGT/RORC INVERSE AGONISTS | | Descriptor: | 2-[(2S)-4-[(4-fluorophenyl)sulfonyl]-7-(1,1,1,3,3,3-hexafluoro-2-hydroxypropan-2-yl)-3,4-dihydro-2H-1,4-benzothiazin-2-yl]-N-(2-hydroxy-2-methylpropyl)acetamide, Nuclear receptor ROR-gamma, SULFATE ION | | Authors: | Sack, J. | | Deposit date: | 2017-11-16 | | Release date: | 2017-12-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of bicyclic hexafluoroisopropyl alcohol sulfonamides as retinoic acid receptor-related orphan receptor gamma (ROR gamma /RORc) inverse agonists. Employing structure-based drug design to improve pregnane X receptor (PXR) selectivity.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
1UHX
 
 | | Crystal structure of d(GCGAGAGC): the base-intercalated duplex | | Descriptor: | 5'-D(*GP*(CBR)P*GP*AP*GP*AP*GP*C)-3', CHLORIDE ION, COBALT HEXAMMINE(III), ... | | Authors: | Kondo, J, Umeda, S.I, Fujita, K, Sunami, T, Takenaka, A. | | Deposit date: | 2003-07-13 | | Release date: | 2004-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray analyses of d(GCGAXAGC) containing G and T at X: the base-intercalated duplex is still stable even in point mutants at the fifth residue.
J.Synchrotron Radiat., 11, 2004
|
|
5A9Z
 
 | | Complex of Thermous thermophilus ribosome bound to BipA-GDPCP | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Kumar, V, Chen, Y, Ahmed, T, Tan, J, Ero, R, Bhushan, S, Gao, Y.-G. | | Deposit date: | 2015-07-23 | | Release date: | 2015-10-14 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structure of Bipa in GTP Form Bound to the Ratcheted Ribosome.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1UI0
 
 | | Crystal Structure Of Uracil-DNA Glycosylase From Thermus Thermophilus HB8 | | Descriptor: | IRON/SULFUR CLUSTER, SULFATE ION, URACIL, ... | | Authors: | Hoseki, J, Okamoto, A, Masui, R, Shibata, T, Inoue, Y, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-14 | | Release date: | 2003-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of a Family 4 Uracil-DNA Glycosylase from Thermus thermophilus HB8
J.Mol.Biol., 333, 2003
|
|
8IJT
 
 | | crystal structure of Hyp N135A mutant from Hypoxylon sp. E7406B | | Descriptor: | Terpene synthase | | Authors: | Gao, J, Su, L.Q, Li, Q, Han, X, Wei, H.L, Dai, Z.J, Liu, W.D. | | Deposit date: | 2023-02-28 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | crystal structure of Hyp N135A mutant from Hypoxylon sp. E7406B
to be published
|
|
5AHZ
 
 | | Bromide-bound form of Halorhodopsin from Halobacterium salinarum in a new rhombohedral crystal form | | Descriptor: | BROMIDE ION, HALORHODOPSIN, RETINAL, ... | | Authors: | Schreiner, M, Schlesinger, R, Heberle, J, Niemann, H.H. | | Deposit date: | 2015-02-11 | | Release date: | 2015-04-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of Halorhodopsin from Halobacterium Salinarum in a New Crystal Form that Imposes Little Restraint on the E-F Loop.
J.Struct.Biol., 190, 2015
|
|
8I5M
 
 | | Rat Kir4.1 in complex with PIP2 | | Descriptor: | ATP-sensitive inward rectifier potassium channel 10, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Zhao, C, Guo, J. | | Deposit date: | 2023-01-26 | | Release date: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Pharmacological inhibition of astrocytic Kir4.1 channel evokes rapid-onset antidepressant responses
To Be Published
|
|
1G06
 
 | | CRYSTAL STRUCTURE OF T4 LYSOZYME MUTANT V149S | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, PROTEIN (LYSOZYME) | | Authors: | Xu, J, Baase, W.A, Quillin, M.L, Matthews, B.W. | | Deposit date: | 2000-10-05 | | Release date: | 2001-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and thermodynamic analysis of the binding of solvent at internal sites in T4 lysozyme.
Protein Sci., 10, 2001
|
|
1I4K
 
 | | CRYSTAL STRUCTURE OF AN SM-LIKE PROTEIN (AF-SM1) FROM ARCHAEOGLOBUS FULGIDUS AT 2.5A RESOLUTION | | Descriptor: | CITRIC ACID, PUTATIVE SNRNP SM-LIKE PROTEIN | | Authors: | Toro, I, Thore, S, Mayer, C, Basquin, J, Seraphin, B, Suck, D. | | Deposit date: | 2001-02-22 | | Release date: | 2001-08-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | RNA binding in an Sm core domain: X-ray structure and functional analysis of an archaeal Sm protein complex.
EMBO J., 20, 2001
|
|
1G0L
 
 | | CRYSTAL STRUCTURE OF T4 LYSOZYME MUTANT T152V | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, PROTEIN (LYSOZYME) | | Authors: | Xu, J, Baase, W.A, Quillin, M.L, Matthews, B.W. | | Deposit date: | 2000-10-06 | | Release date: | 2001-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and thermodynamic analysis of the binding of solvent at internal sites in T4 lysozyme.
Protein Sci., 10, 2001
|
|
2ZQS
 
 | |
5AJO
 
 | | Crystal structure of the inactive form of GalNAc-T2 in complex with the glycopeptide MUC5AC-3,13 | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, MUCIN, POLYPEPTIDE N-ACETYLGALACTOSAMINYLTRANSFERASE 2, ... | | Authors: | Lira-Navarrete, E, delasRivas, M, Companon, I, Pallares, M.C, Kong, Y, Iglesias-Fernandez, J, Bernardes, G.J.L, Peregrina, J.M, Rovira, C, Bernado, P, Bruscolini, P, Clausen, H, Lostao, A, Corzana, F, Hurtado-Guerrero, R. | | Deposit date: | 2015-02-26 | | Release date: | 2015-03-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Dynamic Interplay between Catalytic and Lectin Domains of Galnac-Transferases Modulates Protein O-Glycosylation.
Nat.Commun., 6, 2015
|
|
3B79
 
 | |
8I5N
 
 | | Rat Kir4.1 in complex with PIP2 and Lys05 | | Descriptor: | ATP-sensitive inward rectifier potassium channel 10, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Zhao, C, Guo, J. | | Deposit date: | 2023-01-26 | | Release date: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Pharmacological inhibition of astrocytic Kir4.1 channel evokes rapid-onset antidepressant responses
To Be Published
|
|
1G07
 
 | | CRYSTAL STRUCTURE OF T4 LYSOZYME MUTANT V149C | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, PROTEIN (LYSOZYME) | | Authors: | Xu, J, Baase, W.A, Quillin, M.L, Matthews, B.W. | | Deposit date: | 2000-10-05 | | Release date: | 2001-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and thermodynamic analysis of the binding of solvent at internal sites in T4 lysozyme.
Protein Sci., 10, 2001
|
|
5AHI
 
 | | Crystal structure of salmonalla enterica HisA mutant D7N with ProFAR | | Descriptor: | 1-(5-PHOSPHORIBOSYL)-5-[(5-PHOSPHORIBOSYLAMINO) METHYLIDENE AMINO] IMIDAZOLE-4-CARBOXAMIDE ISOMERASE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Soderholm, A, Guo, X, Newton, M.S, Evans, G.B, Nasvall, J, Patrick, W.M, Selmer, M. | | Deposit date: | 2015-02-06 | | Release date: | 2016-03-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Structure and Mechanism of Hisa from Salmonella Enterica
To be Published
|
|
8AOV
 
 | |
3J29
 
 | | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM | | Descriptor: | 16S rRNA | | Authors: | Guo, Q, Goto, S, Chen, Y, Muto, A, Himeno, H, Deng, H, Lei, J, Gao, N. | | Deposit date: | 2012-09-28 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (14 Å) | | Cite: | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM and general features of the assembly process
Nucleic Acids Res., 41, 2013
|
|
8ACT
 
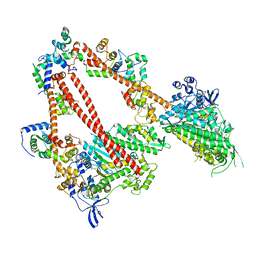 | | structure of the human beta-cardiac myosin folded-back off state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Myosin light chain 3, ... | | Authors: | Grinzato, A, Kandiah, E, Robert-Paganin, J, Auguin, D, Kikuti, C, Nandwani, N, Moussaoui, D, Pathak, D, Ruppel, K.M, Spudich, J.A, Houdusse, A. | | Deposit date: | 2022-07-06 | | Release date: | 2023-06-07 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of the folded-back state of human beta-cardiac myosin.
Nat Commun, 14, 2023
|
|
5A31
 
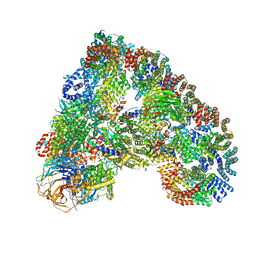 | | Structure of the human APC-Cdh1-Hsl1-UbcH10 complex. | | Descriptor: | ANAPHASE-PROMOTING COMPLEX SUBUNIT 1, ANAPHASE-PROMOTING COMPLEX SUBUNIT 10, ANAPHASE-PROMOTING COMPLEX SUBUNIT 11, ... | | Authors: | Chang, L, Zhang, Z, Yang, J, Mclaughlin, S.H, Barford, D. | | Deposit date: | 2015-05-26 | | Release date: | 2015-11-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Atomic Structure of the Apc/C and its Mechanism of Protein Ubiquitination.
Nature, 522, 2015
|
|
8J2K
 
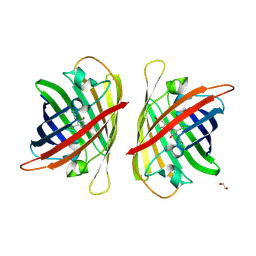 | | Crystal structure of a bright green fluorescent protein (StayGold) with double mutation (N137A, Q140S) in jellyfish Cytaeis uchidae from Biortus | | Descriptor: | 1,2-ETHANEDIOL, StayGold(N137A, Q140S) | | Authors: | Wu, J, Wang, F, Gui, W, Cheng, W, Yang, Y. | | Deposit date: | 2023-04-14 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a bright green fluorescent protein (StayGold) in jellyfish Cytaeis uchidae from Biortus
To Be Published
|
|
3B96
 
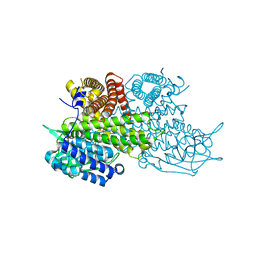 | | Structural Basis for Substrate Fatty-Acyl Chain Specificity: Crystal Structure of Human Very-Long-Chain Acyl-CoA Dehydrogenase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, TETRADECANOYL-COA, Very long-chain specific acyl-CoA dehydrogenase | | Authors: | McAndrew, R.P, Wang, Y, Mohsen, A.W, He, M, Vockley, J, Kim, J.J. | | Deposit date: | 2007-11-02 | | Release date: | 2008-02-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural basis for substrate fatty acyl chain specificity: crystal structure of human very-long-chain acyl-CoA dehydrogenase.
J.Biol.Chem., 283, 2008
|
|
