8DTM
 
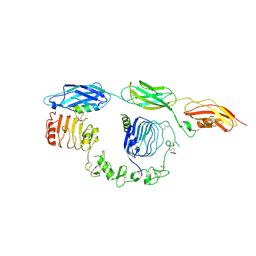 | | Cryo-EM structure of insulin receptor (IR) bound with S597 component 2 | | Descriptor: | Insulin mimetic peptide S597 component 2, Insulin receptor | | Authors: | Park, J, Li, J, Mayer, J.P, Ball, K.A, Wu, J.Y, Hall, C, Accili, D, Stowell, M.H.B, Bai, X.C, Choi, E. | | Deposit date: | 2022-07-26 | | Release date: | 2022-09-07 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Activation of the insulin receptor by an insulin mimetic peptide.
Nat Commun, 13, 2022
|
|
8DTL
 
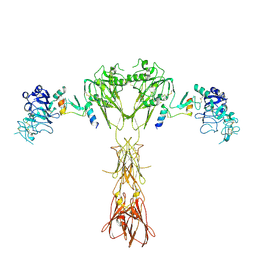 | | Cryo-EM structure of insulin receptor (IR) bound with S597 peptide | | Descriptor: | Insulin mimetic peptide S597, Insulin receptor | | Authors: | Park, J, Li, J, Mayer, J.P, Ball, K.A, Wu, J.Y, Hall, C, Accili, D, Stowell, M.H.B, Bai, X.C, Choi, E. | | Deposit date: | 2022-07-25 | | Release date: | 2022-09-07 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Activation of the insulin receptor by an insulin mimetic peptide.
Nat Commun, 13, 2022
|
|
8DKN
 
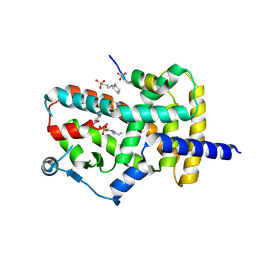 | | PPARg bound to T0070907 and Co-R peptide | | Descriptor: | 2-chloro-5-nitro-N-(pyridin-4-yl)benzamide, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, Nuclear receptor corepressor 1 peptide, ... | | Authors: | Larsen, N.A, Tsai, J. | | Deposit date: | 2022-07-05 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Biochemical and structural basis for the pharmacological inhibition of nuclear hormone receptor PPAR gamma by inverse agonists.
J.Biol.Chem., 298, 2022
|
|
8GOA
 
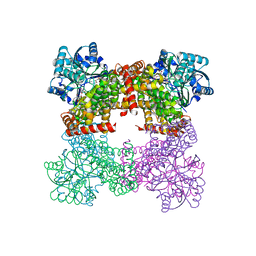 | | Crystal Structure of Glycerol Dehydrogenase in the absence of NAD+ | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glycerol dehydrogenase, ZINC ION | | Authors: | Park, T, Hoang, H.N, Kang, J.Y, Park, J, Mun, S.A, Jin, M, Yang, J, Jung, C.-H, Eom, S.H. | | Deposit date: | 2022-08-24 | | Release date: | 2023-06-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and functional insights into the flexible beta-hairpin of glycerol dehydrogenase.
Febs J., 290, 2023
|
|
5J7A
 
 | | Bacteriorhodopsin ground state structure obtained with Serial Femtosecond Crystallography | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, RETINAL | | Authors: | Nogly, P, Panneels, V, Nelson, G, Gati, C, Kimura, T, Milne, C, Milathianaki, D, Kubo, M, Wu, W, Conrad, C, Coe, J, Bean, R, Zhao, Y, Bath, P, Dods, R, Harimoorthy, R, Beyerlein, K.R, Rheinberger, J, James, D, DePonte, D, Li, C, Sala, L, Williams, G, Hunter, M, Koglin, J.E, Berntsen, P, Nango, E, Iwata, S, Chapman, H.N, Fromme, P, Frank, M, Abela, R, Boutet, S, Barty, A, White, T.A, Weierstall, U, Spence, J, Neutze, R, Schertler, G, Standfuss, J. | | Deposit date: | 2016-04-06 | | Release date: | 2016-08-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Lipidic cubic phase injector is a viable crystal delivery system for time-resolved serial crystallography.
Nat Commun, 7, 2016
|
|
6XP6
 
 | | 3C11-DQ2-glia-a2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3.C11 IgH Fab, ... | | Authors: | Petersen, J, Rossjohn, J. | | Deposit date: | 2020-07-08 | | Release date: | 2021-07-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A high-affinity human TCR-like antibody detects celiac disease gluten peptide-MHC complexes and inhibits T cell activation.
Sci Immunol, 6, 2021
|
|
8KD5
 
 | | Rpd3S in complex with nucleosome with H3K36MLA modification and 187bp DNA, class2 | | Descriptor: | 187bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Chang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
6PIB
 
 | | Structure of the Klebsiella pneumoniae LpxH-AZ1 complex | | Descriptor: | 1-[5-({4-[3-(trifluoromethyl)phenyl]piperazin-1-yl}sulfonyl)-2,3-dihydro-1H-indol-1-yl]ethan-1-one, MANGANESE (II) ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Cho, J, Zhou, P. | | Deposit date: | 2019-06-26 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural basis of the UDP-diacylglucosamine pyrophosphohydrolase LpxH inhibition by sulfonyl piperazine antibiotics.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8KD3
 
 | | Rpd3S in complex with nucleosome with H3K36MLA modification, H3K9Q mutation and 187bp DNA | | Descriptor: | 187bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Zhang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
8KD6
 
 | | Rpd3S in complex with nucleosome with H3K36MLA modification and 187bp DNA, class3 | | Descriptor: | 187bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Zhang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
6XNG
 
 | | MHC-like protein complex structure | | Descriptor: | (3R)-N-[(2S,3R)-1-(alpha-D-galactopyranosyloxy)-3-hydroxyheptadecan-2-yl]-3-hydroxyheptadecanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Thirunavukkarasu, P, Le Nours, J, Rossjohn, J. | | Deposit date: | 2020-07-02 | | Release date: | 2021-11-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Host immunomodulatory lipids created by symbionts from dietary amino acids.
Nature, 600, 2021
|
|
8KC7
 
 | | Rpd3S histone deacetylase complex | | Descriptor: | Chromatin modification-related protein EAF3, Histone deacetylase RPD3, Transcriptional regulatory protein RCO1, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Zhang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-06 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
8KD4
 
 | | Rpd3S in complex with nucleosome with H3K36MLA modification and 187bp DNA, class1 | | Descriptor: | 187bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Zhang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
8KD2
 
 | | Rpd3S in complex with 187bp nucleosome | | Descriptor: | 187bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Zhang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
8AAE
 
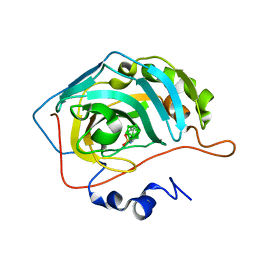 | | CAII in complex with para-carboran-propylsulfonamid | | Descriptor: | Carbonic anhydrase 2, Para-Carborane propyl-sulfonamide, ZINC ION | | Authors: | Brynda, J, Rezacova, P. | | Deposit date: | 2022-07-01 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | B-H⋯ pi and C-H⋯ pi interactions in protein-ligand complexes: carbonic anhydrase II inhibition by carborane sulfonamides.
Phys Chem Chem Phys, 25, 2023
|
|
8KD7
 
 | | Rpd3S in complex with nucleosome with H3K36MLA modification and 167bp DNA | | Descriptor: | 167bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Chang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
8AA6
 
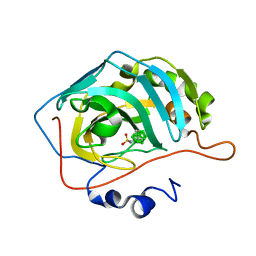 | | CAII in complex with meta-carboran-propylsulfonamid | | Descriptor: | Carbonic anhydrase 2, ZINC ION, meta-carboran-propylsulfonamid | | Authors: | Brynda, J, Rezacova, P, Kugler, M. | | Deposit date: | 2022-06-30 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | B-H⋯ pi and C-H⋯ pi interactions in protein-ligand complexes: carbonic anhydrase II inhibition by carborane sulfonamides.
Phys Chem Chem Phys, 25, 2023
|
|
8AUV
 
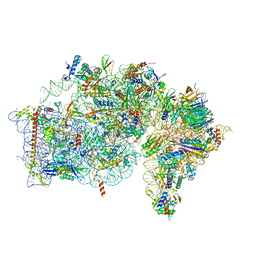 | | Cryo-EM structure of the plant 40S subunit | | Descriptor: | 18S rRNA, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Smirnova, J, Loerke, J, Kleinau, G, Schmidt, A, Buerger, J, Meyer, E.H, Mielke, T, Scheerer, P, Bock, R, Spahn, C.M.T, Zoschke, R. | | Deposit date: | 2022-08-25 | | Release date: | 2023-06-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.38 Å) | | Cite: | Structure of the actively translating plant 80S ribosome at 2.2 angstrom resolution.
Nat.Plants, 9, 2023
|
|
5F81
 
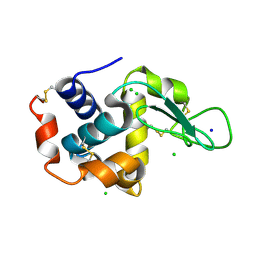 | | Acoustic injectors for drop-on-demand serial femtosecond crystallography | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Roessler, C.G, Agarwal, R, Allaire, M, Alonso-Mori, R, Andi, B, Bachega, J.F.R, Bommer, M, Brewster, A.S, Browne, M.C, Chatterjee, R, Cho, E, Cohen, A.E, Cowan, M, Datwani, S, Davidson, V.L, Defever, J, Eaton, B, Ellson, R, Feng, Y, Ghislain, L.P, Glownia, J.M, Han, G, Hattne, J, Hellmich, J, Heroux, A, Ibrahim, M, Kern, J, Kuczewski, A, Lemke, H.T, Liu, P, Majlof, L, McClintock, W.M, Myers, S, Nelsen, S, Olechno, J, Orville, A.M, Sauter, N.K, Soares, A.S, Soltis, M.S, Song, H, Stearns, R.G, Tran, R, Tsai, Y, Uervirojnangkoorn, M, Wilmot, C.M, Yachandra, V, Yano, J, Yukl, E.T, Zhu, D, Zouni, A. | | Deposit date: | 2015-12-08 | | Release date: | 2016-02-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Acoustic Injectors for Drop-On-Demand Serial Femtosecond Crystallography.
Structure, 24, 2016
|
|
6YLT
 
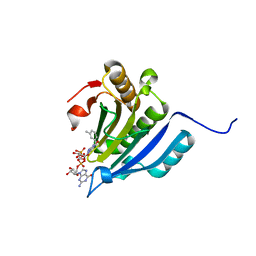 | | Translation initiation factor 4E in complex with 3-MeBn7GpppG mRNA 5' cap analog | | Descriptor: | Eukaryotic translation initiation factor 4E, [[(2~{R},3~{S},4~{R},5~{R})-5-[2-azanyl-7-[(3-methylphenyl)methyl]-6-oxidanylidene-1~{H}-purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [[(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-6-oxidanylidene-1~{H}-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] hydrogen phosphate | | Authors: | Kubacka, D, Wojcik, R, Baranowski, M.R, Kowalska, J, Jemielity, J. | | Deposit date: | 2020-04-07 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Novel N7-Arylmethyl Substituted Dinucleotide mRNA 5' cap Analogs: Synthesis and Evaluation as Modulators of Translation.
Pharmaceutics, 13, 2021
|
|
4TNJ
 
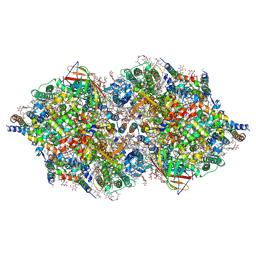 | | RT XFEL structure of Photosystem II 500 ms after the 2nd illumination (2F) at 4.5 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Tran, R, Alonso-Mori, R, Koroidov, S, Echols, N, Hattne, J, Ibrahim, M, Gul, S, Laksmono, H, Sierra, R.G, Gildea, R.J, Han, G, Hellmich, J, Lassalle-Kaiser, B, Chatterjee, R, Brewster, A, Stan, C.A, Gloeckner, C, Lampe, A, DiFiore, D, Milathianaki, D, Fry, A.R, Seibert, M.M, Koglin, J.E, Gallo, E, Uhlig, J, Sokaras, D, Weng, T.-C, Zwart, P.H, Skinner, D.E, Bogan, M.J, Messerschmidt, M, Glatzel, P, Williams, G.J, Boutet, S, Adams, P.D, Zouni, A, Messinger, J, Sauter, N.K, Bergmann, U, Yano, J, Yachandra, V.K. | | Deposit date: | 2014-06-04 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Taking snapshots of photosynthetic water oxidation using femtosecond X-ray diffraction and spectroscopy.
Nat Commun, 5, 2014
|
|
1DGI
 
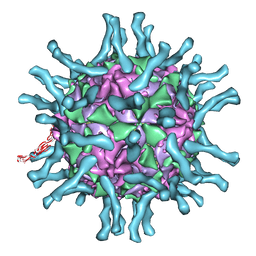 | | Cryo-EM structure of human poliovirus(serotype 1)complexed with three domain CD155 | | Descriptor: | POLIOVIRUS RECEPTOR, VP1, VP2, ... | | Authors: | He, Y, Bowman, V.D, Mueller, S, Bator, C.M, Bella, J, Peng, X, Baker, T.S, Wimmer, E, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 1999-11-24 | | Release date: | 2000-01-24 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (22 Å) | | Cite: | Interaction of the poliovirus receptor with poliovirus.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
4TNI
 
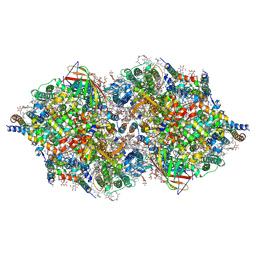 | | RT XFEL structure of Photosystem II 500 ms after the third illumination at 4.6 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Tran, R, Alonso-Mori, R, Koroidov, S, Echols, N, Hattne, J, Ibrahim, M, Gul, S, Laksmono, H, Sierra, R.G, Gildea, R.J, Han, G, Hellmich, J, Lassalle-Kaiser, B, Chatterjee, R, Brewster, A, Stan, C.A, Gloeckner, C, Lampe, A, DiFiore, D, Milathianaki, D, Fry, A.R, Seibert, M.M, Koglin, J.E, Gallo, E, Uhlig, J, Sokaras, D, Weng, T.-C, Zwart, P.H, Skinner, D.E, Bogan, M.J, Messerschmidt, M, Glatzel, P, Williams, G.J, Boutet, S, Adams, P.D, Zouni, A, Messinger, J, Sauter, N.K, Bergmann, U, Yano, J, Yachandra, V.K. | | Deposit date: | 2014-06-04 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (4.6 Å) | | Cite: | Taking snapshots of photosynthetic water oxidation using femtosecond X-ray diffraction and spectroscopy.
Nat Commun, 5, 2014
|
|
6YLR
 
 | | Translation initiation factor 4E in complex with bn7GpppG mRNA 5' cap analog | | Descriptor: | Eukaryotic translation initiation factor 4E, bn7GpppG mRNA 5' cap analog | | Authors: | Kubacka, D, Wojcik, R, Baranowski, M.R, Warminski, M, Kowalska, J, Jemielity, J. | | Deposit date: | 2020-04-07 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1954546 Å) | | Cite: | Novel N7-Arylmethyl Substituted Dinucleotide mRNA 5' cap Analogs: Synthesis and Evaluation as Modulators of Translation.
Pharmaceutics, 13, 2021
|
|
5J2S
 
 | | NKR-P1B from Rattus norvegicus | | Descriptor: | Killer cell lectin-like receptor subfamily B member 1B allele A, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Skalova, T, Vanek, O, Blaha, J, Duskova, J, Hasek, J, Koval, T, Dohnalek, J. | | Deposit date: | 2016-03-30 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of NKR-P1B from Rattus norvegicus
To Be Published
|
|
