2YKN
 
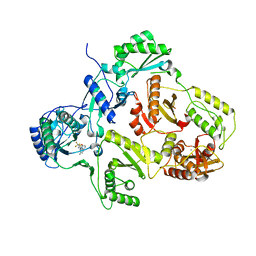 | | Crystal structure of HIV-1 Reverse Transcriptase (RT) in complex with a Difluoromethylbenzoxazole (DFMB) Pyrimidine Thioether derivative, a non-nucleoside RT inhibitor (NNRTI) | | Descriptor: | 2-[DIFLUORO-[(4-METHYL-PYRIMIDINYL)-THIO]METHYL]-BENZOXAZOLE, CALCIUM ION, REVERSE TRANSCRIPTASE/RIBONUCLEASE H | | Authors: | Boyer, J, Arnoult, E, Medebielle, M, Guillemont, J, Unge, T, Unge, J, Jochmans, D. | | Deposit date: | 2011-05-28 | | Release date: | 2011-08-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Difluoromethylbenzoxazole Pyrimidine Thioether Derivatives: A Novel Class of Potent Non-Nucleoside HIV-1 Reverse Transcriptase Inhibitors.
J.Med.Chem., 54, 2011
|
|
4Q1B
 
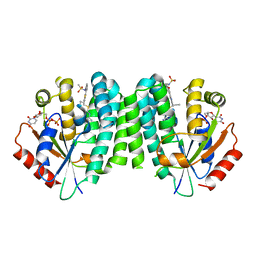 | |
6TQU
 
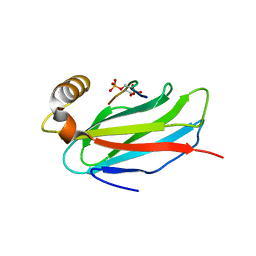 | | The crystal structure of the MSP domain of human MOSPD2 in complex with the Phospho-FFAT motif of STARD3. | | Descriptor: | Motile sperm domain-containing protein 2, SULFATE ION, StAR-related lipid transfer protein 3 | | Authors: | McEwen, A.G, Poussin-Courmontagne, P, Di Mattia, T, Wendling, C, Cavarelli, J, Tomasetto, C, Alpy, F. | | Deposit date: | 2019-12-17 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | FFAT motif phosphorylation controls formation and lipid transfer function of inter-organelle contacts.
Embo J., 39, 2020
|
|
6THC
 
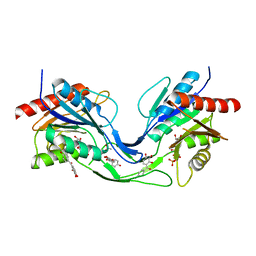 | | Crystal structure of Mycobacterium smegmatis CoaB in complex with CTP and (4-hydroxyphenyl)(2,3,4-trihydroxyphenyl)methanone | | Descriptor: | (4-hydroxyphenyl)-[2,3,4-tris(oxidanyl)phenyl]methanone, ACETATE ION, CALCIUM ION, ... | | Authors: | Mendes, V, Blaszczyk, M, Bryant, O, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2019-11-19 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.033 Å) | | Cite: | Inhibiting Mycobacterium tuberculosis CoaBC by targeting an allosteric site.
Nat Commun, 12, 2021
|
|
4Q19
 
 | | Human dCK C4S-S74E mutant in complex with UDP and the inhibitor 5 {5-(4-{[(4,6-DIAMINOPYRIMIDIN-2-YL)SULFANYL]METHYL}-5-PROPYL-1,3-THIAZOL-2-YL)-2-METHOXYPHENOL} | | Descriptor: | 5-(4-(((4,6-diaminopyrimidin-2-yl)thio)methyl)-5-propylthiazol-2-yl)-2-methoxyphenol, Deoxycytidine kinase, URIDINE-5'-DIPHOSPHATE | | Authors: | Nomme, J, Lavie, A. | | Deposit date: | 2014-04-03 | | Release date: | 2014-11-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure-guided development of deoxycytidine kinase inhibitors with nanomolar affinity and improved metabolic stability.
J.Med.Chem., 57, 2014
|
|
6TM0
 
 | | N-Domain P40/P90 Mycoplasma pneumoniae complexed with 6'SL | | Descriptor: | Mgp-operon protein 3, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Vizarraga, D, Aparicio, D, Illanes, R, Fita, I, Perez-Luque, R, Martin, J. | | Deposit date: | 2019-12-03 | | Release date: | 2020-11-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Immunodominant proteins P1 and P40/P90 from human pathogen Mycoplasma pneumoniae.
Nat Commun, 11, 2020
|
|
5M5F
 
 | | Thermolysin in complex with inhibitor and krypton | | Descriptor: | (2~{S})-4-methyl-2-[2-[[oxidanyl(phenylmethoxycarbonylaminomethyl)phosphoryl]amino]ethanoylamino]pentanoic acid, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Krimmer, S.G, Cramer, J, Heine, A, Klebe, G. | | Deposit date: | 2016-10-21 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | How Nothing Boosts Affinity: Hydrophobic Ligand Binding to the Virtually Vacated S1' Pocket of Thermolysin.
J. Am. Chem. Soc., 139, 2017
|
|
8TFN
 
 | | Structure of anti-TCRvbeta6-5 antibody in complex with the cognate TCR | | Descriptor: | Anti-TCRVb6-5 Fab heavy chain, Anti-TCRVb6-5 Fab light chain, TRAV12-3, ... | | Authors: | Katragadda, M, Servatalab, R, Wirth, J. | | Deposit date: | 2023-07-11 | | Release date: | 2023-11-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Innate TCR beta-chain engagement drives human T cells toward distinct memory-like effector phenotypes with immunotherapeutic potentials.
Sci Adv, 9, 2023
|
|
6TPA
 
 | | CDK8/CyclinC in complex with drug ETP-50775 | | Descriptor: | (2~{R})-butane-1,2-diol, 1-[4-chloranyl-3-(trifluoromethyl)phenyl]-3-(5-oxidanylidene-6-pyridin-4-yl-pyrido[2,3-b][1,5]benzoxazepin-9-yl)urea, Cyclin-C, ... | | Authors: | Munoz, I.G, Pastor, J, Martinez, S. | | Deposit date: | 2019-12-12 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Pyrido[2,3-b][1,5]benzoxazepin-5(6H)-one derivatives as CDK8 inhibitors.
Eur.J.Med.Chem., 201, 2020
|
|
7YP0
 
 | | Crystal structure of CtGST | | Descriptor: | Glutathione S-transferase | | Authors: | Yang, J, Fan, J.P, Lei, X.G. | | Deposit date: | 2022-08-02 | | Release date: | 2024-02-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Enzymatic Degradation of Deoxynivalenol with the Engineered Detoxification Enzyme Fhb7.
Jacs Au, 4, 2024
|
|
7YOC
 
 | | Crystal structure of Fhb7 | | Descriptor: | Fhb7 | | Authors: | Yang, J, Liang, K, Xiao, J.Y, Lei, X.G. | | Deposit date: | 2022-08-01 | | Release date: | 2024-02-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Enzymatic Degradation of Deoxynivalenol with the Engineered Detoxification Enzyme Fhb7.
Jacs Au, 4, 2024
|
|
7LHT
 
 | | Structure of the LRRK2 dimer | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, Leucine-rich repeat serine/threonine-protein kinase 2 | | Authors: | Myasnikov, A, Zhu, H, Hixson, P, Xie, B, Yu, K, Pitre, A, Peng, J, Sun, J. | | Deposit date: | 2021-01-26 | | Release date: | 2021-06-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural analysis of the full-length human LRRK2.
Cell, 184, 2021
|
|
7LHW
 
 | | Structure of the LRRK2 monomer | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, Leucine-rich repeat serine/threonine-protein kinase 2 | | Authors: | Myasnikov, A, Zhu, H, Hixson, P, Xie, B, Yu, K, Pitre, A, Peng, J, Sun, J. | | Deposit date: | 2021-01-26 | | Release date: | 2021-06-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural analysis of the full-length human LRRK2.
Cell, 184, 2021
|
|
7LI3
 
 | | Structure of the LRRK2 G2019S mutant | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, Leucine-rich repeat serine/threonine-protein kinase 2 | | Authors: | Myasnikov, A, Zhu, H, Hixson, P, Xie, B, Yu, K, Pitre, A, Peng, J, Sun, J. | | Deposit date: | 2021-01-26 | | Release date: | 2021-06-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural analysis of the full-length human LRRK2.
Cell, 184, 2021
|
|
4Q18
 
 | | Human dCK C4S-S74E mutant in complex with UDP and the inhibitor 4 [1-[5-(4-{[(2,6-diaminopyrimidin-4-yl)sulfanyl]methyl}-5-propyl-1,3-thiazol-2-yl)-2-methoxyphenoxy]-2-methylpropan-2-ol] | | Descriptor: | 1-(5-(4-(((2,6-diaminopyrimidin-4-yl)thio)methyl)-5-propylthiazol-2-yl)-2-methoxyphenoxy)-2-methylpropan-2-ol, Deoxycytidine kinase, URIDINE-5'-DIPHOSPHATE | | Authors: | Nomme, J, Lavie, A. | | Deposit date: | 2014-04-03 | | Release date: | 2014-11-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-guided development of deoxycytidine kinase inhibitors with nanomolar affinity and improved metabolic stability.
J.Med.Chem., 57, 2014
|
|
7LI4
 
 | | Structure of LRRK2 after symmetry expansion | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, Leucine-rich repeat serine/threonine-protein kinase 2 | | Authors: | Myasnikov, A, Zhu, H, Hixson, P, Xie, B, Yu, K, Pitre, A, Peng, J, Sun, J. | | Deposit date: | 2021-01-26 | | Release date: | 2021-06-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural analysis of the full-length human LRRK2.
Cell, 184, 2021
|
|
7ZR9
 
 | | OMI-2 FAB IN COMPLEX WITH SARS-COV-2 BETA SPIKE GLYCOPROTEIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Omi-2 Fab heavy chain, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | Deposit date: | 2022-05-03 | | Release date: | 2022-06-01 | | Last modified: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
7ZRC
 
 | | OMI-38 FAB IN COMPLEX WITH SARS-COV-2 BETA SPIKE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Omi-38 Fab Heavy Chain, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | Deposit date: | 2022-05-04 | | Release date: | 2022-06-01 | | Last modified: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
5LZT
 
 | | Structure of the mammalian ribosomal termination complex with eRF1 and eRF3. | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Shao, S, Murray, J, Brown, A, Taunton, J, Ramakrishnan, V, Hegde, R.S. | | Deposit date: | 2016-10-02 | | Release date: | 2016-11-30 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Decoding Mammalian Ribosome-mRNA States by Translational GTPase Complexes.
Cell, 167, 2016
|
|
4WWO
 
 | | Crystal structure of human PI3K-gamma in complex with phenylquinoline inhibitor N-{(1S)-1-[8-chloro-2-(3-fluorophenyl)quinolin-3-yl]ethyl}-9H-purin-6-amine | | Descriptor: | N-{(1S)-1-[8-chloro-2-(3-fluorophenyl)quinolin-3-yl]ethyl}-9H-purin-6-amine, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Whittington, D.A, Tang, J, Yakowec, P. | | Deposit date: | 2014-11-11 | | Release date: | 2014-12-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery and in Vivo Evaluation of (S)-N-(1-(7-Fluoro-2-(pyridin-2-yl)quinolin-3-yl)ethyl)-9H-purin-6-amine (AMG319) and Related PI3K delta Inhibitors for Inflammation and Autoimmune Disease.
J.Med.Chem., 58, 2015
|
|
7ZR7
 
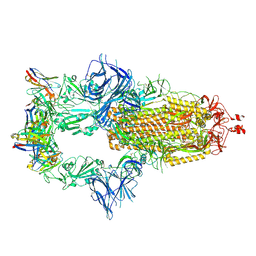 | | OMI-42 FAB IN COMPLEX WITH SARS-COV-2 BETA SPIKE GLYCOPROTEIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Omi-42 heavy chain, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | Deposit date: | 2022-05-03 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
7ZR8
 
 | | OMI-38 FAB IN COMPLEX WITH SARS-COV-2 BETA SPIKE RBD (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Omi-38 Fab light chain, Omi-38 fab heavy chain, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | Deposit date: | 2022-05-03 | | Release date: | 2022-06-01 | | Last modified: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
7LFM
 
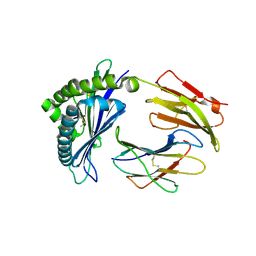 | | MODEL OF MHC CLASS Ib H2-M3 WITH MOUSE ND1 N-TERMINAL HEPTAPEPTIDE, VAL MUTANT, TRICLINIC CELL, REFINED AT 1.60 ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, Heptapeptide from NADH-ubiquinone oxidoreductase chain 1, ... | | Authors: | Tomchick, D.R, Deisenhofer, J, Shen, S. | | Deposit date: | 2021-01-17 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and dynamics of major histocompatibility class Ib molecule H2-M3 complexed with mitochondrial-derived peptides.
J.Biomol.Struct.Dyn., 40, 2022
|
|
7LFL
 
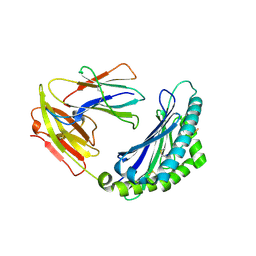 | | MODEL OF MHC CLASS Ib H2-M3 WITH MOUSE ND1 N-TERMINAL HEPTAPEPTIDE, VAL MUTANT, MONOCLINIC CELL, REFINED AT 1.60 ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, Heptapeptide from NADH-ubiquinone oxidoreductase chain 1, ... | | Authors: | Tomchick, D.R, Deisenhofer, J, Shen, S. | | Deposit date: | 2021-01-17 | | Release date: | 2021-07-14 | | Last modified: | 2022-12-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and dynamics of major histocompatibility class Ib molecule H2-M3 complexed with mitochondrial-derived peptides.
J.Biomol.Struct.Dyn., 40, 2022
|
|
8PC7
 
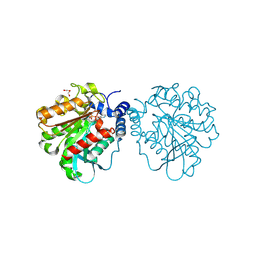 | |
