3MME
 
 | | Structure and functional dissection of PG16, an antibody with broad and potent neutralization of HIV-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PG16 HEAVY CHAIN FAB, ... | | Authors: | Pancera, M, McLellan, J, Zhou, T, Zhu, J, Kwong, P. | | Deposit date: | 2010-04-19 | | Release date: | 2010-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.97 Å) | | Cite: | Crystal structure of PG16 and chimeric dissection with somatically related PG9: structure-function analysis of two quaternary-specific antibodies that effectively neutralize HIV-1.
J.Virol., 84, 2010
|
|
7KC4
 
 | | Human WLS in complex with WNT8A | | Descriptor: | 1-CIS-9-OCTADECANOYL-2-CIS-9-HEXADECANOYL PHOSPHATIDYL GLYCEROL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Nygaard, R, Jia, Y, Kim, J, Ross, D, Parisi, G, Clarke, O.B, Virshup, D.M, Mancia, F. | | Deposit date: | 2020-10-05 | | Release date: | 2021-01-06 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structural Basis of WLS/Evi-Mediated Wnt Transport and Secretion.
Cell, 184, 2021
|
|
3MO8
 
 | | PWWP Domain of Human Bromodomain and PHD finger-containing protein 1 In Complex with Trimethylated H3K36 Peptide | | Descriptor: | Histone H3.2 TRIMETHYLATED H3K36 PEPTIDE, Peregrin | | Authors: | Lam, R, Zeng, H, Ni, S, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-04-22 | | Release date: | 2010-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural and Histone Binding Ability Characterizations of Human PWWP Domains.
Plos One, 6, 2011
|
|
2VM6
 
 | | Human Bcl2-A1 in complex with Bim-BH3 peptide | | Descriptor: | BCL-2-LIKE PROTEIN 11, BCL-2-RELATED PROTEIN A1, SULFATE ION | | Authors: | Herman, M.D, Lehtio, L, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Moche, M, Nilsson, M.E, Nyman, T, Persson, C, Sagemark, J, Svensson, L, Thorsell, A.G, Tresaugues, L, van den Berg, S, Weigelt, J, Welin, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-01-23 | | Release date: | 2008-02-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Completing the Family Portrait of the Anti- Apoptotic Bcl-2 Proteins: Crystal Structure of Human Bfl-1 in Complex with Bim.
FEBS Lett., 582, 2008
|
|
2V4R
 
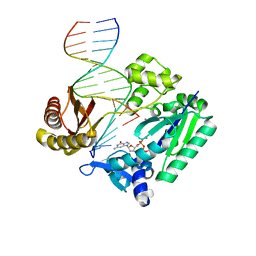 | | Non-productive complex of the Y-family DNA polymerase Dpo4 with dGTP skipping the M1dG adduct to pair with the next template cytosine | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*TP*TP*C)-3', 5'-D(*TP*CP*AP*CP*M1GP*GP*AP*AP*TP*CP*CP *TP*TP*CP*CP*CP*CP*C)-3', ... | | Authors: | Eoff, R.L, Stafford, J.B, Szekely, J, Rizzo, C.J, Egli, M, Guengerich, F.P, Marnett, L.J. | | Deposit date: | 2008-09-26 | | Release date: | 2009-06-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Functional Analysis of Sulfolobus Solfataricus Y-Family DNA Polymerase Dpo4-Catalyzed Bypass of the Malondialdehyde-Deoxyguanosine Adduct.
Biochemistry, 48, 2009
|
|
6I0X
 
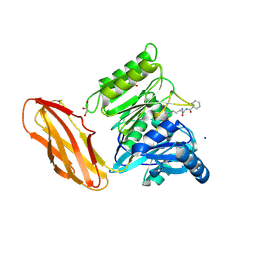 | | Porphyromonas gingivalis peptidylarginine deminase (PPAD) mutant G231N/E232T/N235D in complex with Cl-amidine. | | Descriptor: | GLYCEROL, N-[(1S)-1-(AMINOCARBONYL)-4-(ETHANIMIDOYLAMINO)BUTYL]BENZAMIDE, Peptidylarginine deiminase, ... | | Authors: | Gomis-Ruth, F.X, Goulas, T, Sola, M, Potempa, J. | | Deposit date: | 2018-10-26 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure, function, and inhibition of a genomic/clinical variant of Porphyromonas gingivalis peptidylarginine deiminase.
Protein Sci., 28, 2019
|
|
3V4P
 
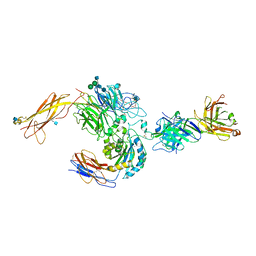 | | crystal structure of a4b7 headpiece complexed with Fab ACT-1 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Yu, Y, Zhu, J, Springer, T.A. | | Deposit date: | 2011-12-15 | | Release date: | 2012-01-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural specializations of a4b7, an Integrin that Mediates Rolling Adhesion
J.Cell Biol., 196, 2012
|
|
9AZB
 
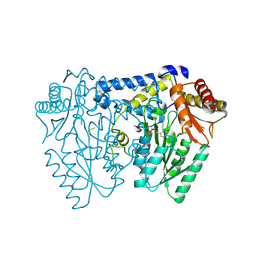 | | Crystal structure of LolTv5 | | Descriptor: | Aminotransferase, class V/Cysteine desulfurase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Gao, J, Hai, Y. | | Deposit date: | 2024-03-11 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Enzymatic Synthesis of Unprotected alpha , beta-Diamino Acids via Direct Asymmetric Mannich Reactions.
J.Am.Chem.Soc., 146, 2024
|
|
5S4G
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with SF005 | | Descriptor: | Non-structural protein 3, SULFATE ION, [1,2,4]triazolo[4,3-a]pyridin-3-amine | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.172 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
2V0P
 
 | | The Structure of Tap42 Alpha4 Subunit | | Descriptor: | TYPE 2A PHOSPHATASE-ASSOCIATED PROTEIN 42, ZINC ION | | Authors: | Roe, S.M, Yang, J, Barford, D. | | Deposit date: | 2007-05-15 | | Release date: | 2007-07-17 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structure of Tap42/Alpha4 Reveals a Tetratricopeptide Repeat-Like Fold and Provides Insights Into Pp2A Regulation.
Biochemistry, 46, 2007
|
|
4GUS
 
 | | Crystal structure of LSD2-NPAC with H3 in space group P3221 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Histone H3.3, ... | | Authors: | Chen, F, Dong, Z, Fang, J, Yang, Y, Li, Z, Xu, Y, Yang, H, Wang, P, Fang, R, Shi, Y, Xu, Y. | | Deposit date: | 2012-08-29 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation
Mol.Cell, 49, 2013
|
|
4GV1
 
 | | PKB alpha in complex with AZD5363 | | Descriptor: | 4-amino-N-[(1S)-1-(4-chlorophenyl)-3-hydroxypropyl]-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidine-4-carboxamide, GLYCEROL, RAC-alpha serine/threonine-protein kinase | | Authors: | Addie, M, Ballard, P, Bird, G, Buttar, D, Currie, G, Davies, B, Debreczeni, J, Dry, H, Dudley, P, Greenwood, R, Hatter, G, Jestel, A, Johnson, P.D, Kettle, J.G, Lane, C, Lamont, G, Leach, A, Luke, R.W.A, Ogilvie, D, Page, K, Pass, M, Steinbacher, S, Steuber, H, Pearson, S, Ruston, L. | | Deposit date: | 2012-08-30 | | Release date: | 2013-02-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Discovery of 4-Amino-N-[(1S)-1-(4-chlorophenyl)-3-hydroxypropyl]-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidine-4-carboxamide (AZD5363), an Orally Bioavailable, Potent Inhibitor of Akt Kinases.
J.Med.Chem., 56, 2013
|
|
2VC7
 
 | | Structural basis for natural lactonase and promiscuous phosphotriesterase activities | | Descriptor: | (4S)-4-(decanoylamino)-5-hydroxy-3,4-dihydro-2H-thiophenium, 1,2-ETHANEDIOL, ARYLDIALKYLPHOSPHATASE, ... | | Authors: | Elias, M, Dupuy, J, Merone, L, Mandrich, L, Moniot, S, Rochu, D, Lecomte, C, Rossi, M, Masson, P, Manco, G, Chabriere, E. | | Deposit date: | 2007-09-19 | | Release date: | 2008-04-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Basis for Natural Lactonase and Promiscuous Phosphotriesterase Activities.
J.Mol.Biol., 379, 2008
|
|
2VEC
 
 | | The crystal structure of the protein YhaK from Escherichia coli | | Descriptor: | CHLORIDE ION, PIRIN-LIKE PROTEIN YHAK | | Authors: | Gurmu, D, Lu, J, Johnson, K.A, Nordlund, P, Holmgren, A, Erlandsen, H. | | Deposit date: | 2007-10-18 | | Release date: | 2008-07-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Crystal Structure of the Protein Yhak from Escherichia Coli Reveals a New Subclass of Redox Sensitive Enterobacterial Bicupins.
Proteins, 74, 2008
|
|
1RU2
 
 | |
2VEX
 
 | | CRYSTAL STRUCTURE OF PROTEIN TYROSINE PHOSPHATASE 1B IN COMPLEX WITH AN ISOTHIAZOLIDINONE-CONTAINING INHIBITOR | | Descriptor: | MAGNESIUM ION, N-{(1S)-2-{4-[(5S)-1,1-dioxido-3-oxoisothiazolidin-5-yl]phenyl}-1-[(4R)-4-(2-phenylethyl)-4,5-dihydro-1H-imidazol-2-yl]ethyl}-3-fluorobenzenesulfonamide, TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE 1 | | Authors: | Douty, B, Wayland, B, Ala, P.J, Bower, M.J, Pruitt, J, Bostrom, L, Wei, M, Klabe, R, Gonneville, L, Wynn, R, Burn, T.C, Liu, P.C.C, Combs, A.P, Yue, E.W. | | Deposit date: | 2007-10-27 | | Release date: | 2007-11-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Isothiazolidinone Inhibitors of Ptp1B Containing Imidazoles and Imidazolines
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2NNC
 
 | | Structure of the sulfur carrier protein SoxY from Chlorobium limicola f thiosulfatophilum | | Descriptor: | CHLORIDE ION, NITROGEN MOLECULE, PHOSPHATE ION, ... | | Authors: | Stout, J, Van Driessche, G, Savvides, S.N, Van Beeumen, J. | | Deposit date: | 2006-10-24 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | X-ray crystallographic analysis of the sulfur carrier protein SoxY from Chlorobium limicola f. thiosulfatophilum reveals a tetrameric structure.
Protein Sci., 16, 2007
|
|
9BER
 
 | | Cryo-EM structure of the HIV-1 JR-FL IDL Env trimer in complex with PGT122 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Gorman, J, Kwong, P.D. | | Deposit date: | 2024-04-16 | | Release date: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Design of soluble HIV-1 envelope trimers free of covalent gp120-gp41 bonds with prevalent native-like conformation.
Cell Rep, 43, 2024
|
|
6I44
 
 | | Allosteric activation of human prekallikrein by apple domain disc rotation | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ... | | Authors: | Li, C, Pathak, M, MaCrae, K, Dreveny, I, Emsley, J. | | Deposit date: | 2018-11-09 | | Release date: | 2019-03-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Plasma kallikrein structure reveals apple domain disc rotated conformation compared to factor XI.
J.Thromb.Haemost., 17, 2019
|
|
4MIJ
 
 | | Crystal structure of a Trap periplasmic solute binding protein from Polaromonas sp. JS666 (Bpro_3107), target EFI-510173, with bound alpha/beta D-Galacturonate, space group P21 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, TRAP dicarboxylate transporter, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Zhao, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-08-31 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
2V8F
 
 | | Mouse Profilin IIa in complex with a double repeat from the FH1 domain of mDia1 | | Descriptor: | GLYCEROL, ISOPROPYL ALCOHOL, PROFILIN-2, ... | | Authors: | Kursula, P, Kursula, I, Downer, J, Witke, W, Wilmanns, M. | | Deposit date: | 2007-08-07 | | Release date: | 2007-12-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | High-Resolution Structural Analysis of Mammalian Profilin 2A Complex Formation with Two Physiological Ligands: The Formin Homology 1 Domain of Mdia1 and the Proline-Rich Domain of Vasp.
J.Mol.Biol., 375, 2008
|
|
1XHX
 
 | | Phi29 DNA Polymerase, orthorhombic crystal form | | Descriptor: | DNA polymerase, MAGNESIUM ION, SULFATE ION | | Authors: | Kamtekar, S, Berman, A.J, Wang, J, Lazaro, J.M, de Vega, M, Blanco, L, Salas, M, Steitz, T.A. | | Deposit date: | 2004-09-21 | | Release date: | 2004-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Insights into Strand Displacement and Processivity from the Crystal Structure of the Protein-Primed DNA Polymerase of Bacteriophage phi29
Mol.Cell, 16, 2004
|
|
2NNF
 
 | | Structure of the sulfur carrier protein SoxY from Chlorobium limicola f thiosulfatophilum | | Descriptor: | PHOSPHATE ION, Sulfur covalently-binding protein | | Authors: | Stout, J, Van Driessche, G, Savvides, S.N, Van Beeumen, J. | | Deposit date: | 2006-10-24 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | X-ray crystallographic analysis of the sulfur carrier protein SoxY from Chlorobium limicola f. thiosulfatophilum reveals a tetrameric structure.
Protein Sci., 16, 2007
|
|
2VDE
 
 | | Crystal Structure of the Open State of TolC Outer Membrane Component of Mutlidrug Efflux Pumps | | Descriptor: | CHLORIDE ION, OUTER MEMBRANE PROTEIN TOLC | | Authors: | Bavro, V.N, Pietras, Z, Furnham, N, Perez-Cano, L, Fernandez-Recio, J, Pei, X.Y, Truer, R, Misra, R, Luisi, B. | | Deposit date: | 2007-10-04 | | Release date: | 2008-04-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Assembly and Channel Opening in a Bacterial Drug Efflux Machine.
Mol.Cell, 30, 2008
|
|
2VK0
 
 | | Crystal structure form ultalente insulin microcrystals | | Descriptor: | 4-HYDROXY-BENZOIC ACID METHYL ESTER, INSULIN A CHAIN, INSULIN B CHAIN, ... | | Authors: | Wagner, A, Diez, J, Schulze-Briese, C, Schluckebier, G. | | Deposit date: | 2007-12-14 | | Release date: | 2008-09-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Ultralente--A Microcrystalline Insulin Suspension.
Proteins, 74, 2009
|
|
