4U7K
 
 | | Crystal structure of the Clostridium histolyticum colH collagenase polycystic kidney disease-like domain 2a in the presence of calcium at 1.9 Angstrom resolution | | Descriptor: | CALCIUM ION, ColH protein | | Authors: | Bauer, R, Janowska, K, Sakon, J, Matsushita, O, Latimer, E. | | Deposit date: | 2014-07-31 | | Release date: | 2015-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structures of three polycystic kidney disease-like domains from Clostridium histolyticum collagenases ColG and ColH.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4UPU
 
 | | Crystal structure of IP3 3-K calmodulin binding region in complex with Calmodulin | | Descriptor: | CALCIUM ION, CALMODULIN, GLYCEROL, ... | | Authors: | Franco-Echevarria, E, Banos-Sanz, J.I, Monterroso, B, Round, A, Sanz-Aparicio, J, Gonzalez, B. | | Deposit date: | 2014-06-18 | | Release date: | 2014-08-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | A New Calmodulin Binding Motif for Inositol 1,4,5-Trisphosphate 3-Kinase Regulation.
Biochem.J., 463, 2014
|
|
4U9B
 
 | | Crystal structure of an H-NOX protein from S. oneidensis in the Fe(II)NO ligation state | | Descriptor: | GLYCEROL, NITRIC OXIDE, NO-binding heme-dependent sensor protein, ... | | Authors: | Herzik Jr, M.A, Jonnalagadda, R, Kuriyan, J, Marletta, M.A. | | Deposit date: | 2014-08-05 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural insights into the role of iron-histidine bond cleavage in nitric oxide-induced activation of H-NOX gas sensor proteins.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4UDX
 
 | | CO2 bound to cluster C of Ni,Fe-CO dehydrogenase at true-atomic resolution | | Descriptor: | CARBON DIOXIDE, CARBON MONOXIDE DEHYDROGENASE 2, FE (II) ION, ... | | Authors: | Fesseler, J, Jeoung, J.-H, Dobbek, H. | | Deposit date: | 2014-12-12 | | Release date: | 2015-05-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | How the [NiFe4S4] Cluster of CO Dehydrogenase Activates CO2 and NCO(-).
Angew. Chem. Int. Ed. Engl., 54, 2015
|
|
4UP5
 
 | | Crystal structure of the Pygo2 PHD finger in complex with the B9L HD1 domain and a chemical fragment | | Descriptor: | 6-methoxy-1,3-benzothiazol-2-amine, PYGOPUS HOMOLOG 2, B-CELL CLL/LYMPHOMA 9-LIKE PROTEIN, ... | | Authors: | Miller, T.C.R, Fiedler, M, Rutherford, T.J, Birchall, K, Chugh, J, Bienz, M. | | Deposit date: | 2014-06-12 | | Release date: | 2014-11-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Competitive Binding of a Benzimidazole to the Histone-Binding Pocket of the Pygo Phd Finger.
Acs Chem.Biol., 9, 2014
|
|
4UNE
 
 | | Human insulin B26Phe mutant crystal structure | | Descriptor: | INSULIN A CHAIN, INSULIN B CHAIN, SULFATE ION | | Authors: | Zakova, L, Klevtikova, E, Lepsik, M, Collinsova, M, Watson, C.J, Turkenburg, J.P, Jiracek, J, Brzozowski, A.M. | | Deposit date: | 2014-05-28 | | Release date: | 2014-10-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Human Insulin Analogues Modified at the B26 Site Reveal a Hormone Conformation that is Undetected in the Receptor Complex
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4UBR
 
 | |
4UIF
 
 | | Cryo-EM structure of Dengue virus serotype 2 in complex with antigen-binding fragments of human antibody 2D22 | | Descriptor: | ANTIGEN-BINDING FRAGMENT OF HUMAN ANTIBODY 2D22 - HEAVY CHAIN, ANTIGEN-BINDING FRAGMENT OF HUMAN ANTIBODY 2D22 - LIGHT CHAIN, DENGUE VIRUS SEROTYPE 2 STRAIN PVP94 07 - ENVELOPE PROTEIN, ... | | Authors: | Fibriansah, G, Ibarra, K.D, Ng, T.-S, Smith, S.A, Tan, J.L, Lim, X.-N, Ooi, J.S.G, Kostyuchenko, V.A, Wang, J, de Silva, A.M, Harris, E, Crowe Junior, J.E, Lok, S.-M. | | Deposit date: | 2015-03-30 | | Release date: | 2015-07-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Cryo-EM structure of an antibody that neutralizes dengue virus type 2 by locking E protein dimers.
Science, 349, 2015
|
|
4UJ6
 
 | | Structure of surface layer protein SbsC, domains 1-6 | | Descriptor: | SURFACE LAYER PROTEIN | | Authors: | Dordic, A, Pavkov-Keller, T, Eder, M, Egelseer, E.M, Davis, K, Mills, D, Sleytr, U.B, Kuehlbrandt, W, Vonck, J, Keller, W. | | Deposit date: | 2015-04-08 | | Release date: | 2016-04-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure of Surface Layer Protein Sbsc, Domains 1-6
To be Published
|
|
3CW8
 
 | | 4-Chlorobenzoyl-CoA Ligase/Synthetase, bound to 4CBA-Adenylate | | Descriptor: | 1,2-ETHANEDIOL, 4-chlorobenzoyl CoA ligase, 5'-O-[(S)-{[(4-chlorophenyl)carbonyl]oxy}(hydroxy)phosphoryl]adenosine | | Authors: | Reger, A.S, Cao, J, Wu, R, Dunaway-Mariano, D, Gulick, A.M. | | Deposit date: | 2008-04-21 | | Release date: | 2008-09-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural characterization of a 140 degrees domain movement in the two-step reaction catalyzed by 4-chlorobenzoate:CoA ligase.
Biochemistry, 47, 2008
|
|
4U7A
 
 | |
4U7U
 
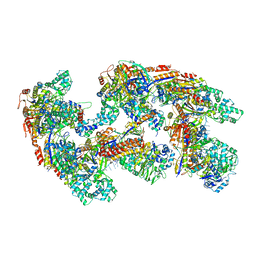 | | Crystal structure of RNA-guided immune Cascade complex from E.coli | | Descriptor: | CRISPR system Cascade subunit CasA, CRISPR system Cascade subunit CasB, CRISPR system Cascade subunit CasC, ... | | Authors: | Zhao, H, Sheng, G, Wang, J, Wang, M, Bunkoczi, G, Gong, W, Wei, Z, Wang, Y. | | Deposit date: | 2014-07-31 | | Release date: | 2014-08-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Crystal structure of the RNA-guided immune surveillance Cascade complex in Escherichia coli
Nature, 515, 2014
|
|
4UBQ
 
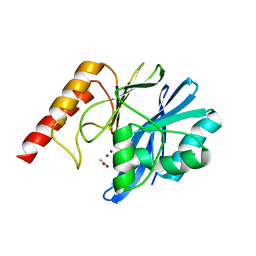 | | Crystal Structure of IMP-2 Metallo-beta-Lactamase from Acinetobacter spp. | | Descriptor: | ACETATE ION, Beta-lactamase, ZINC ION | | Authors: | Yamaguchi, Y, Matsueda, S, Matsunaga, K, Takashio, N, Toma-Fukai, S, Yamagata, Y, Shibata, N, Wachino, J, Shibayama, K, Arakawa, Y, Kurosaki, H. | | Deposit date: | 2014-08-13 | | Release date: | 2014-12-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of IMP-2 metallo-beta-lactamase from Acinetobacter spp.: comparison of active-site loop structures between IMP-1 and IMP-2.
Biol.Pharm.Bull., 38, 2015
|
|
4UC2
 
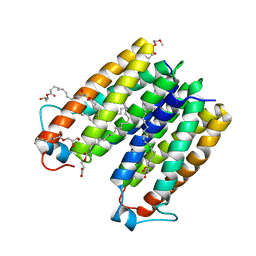 | | Crystal structure of translocator protein 18kDa (TSPO) from rhodobacter sphaeroides (A139T mutant) in P212121 space group | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, TETRAETHYLENE GLYCOL, TRANSLOCATOR PROTEIN TSPO | | Authors: | Li, F, Liu, J, Zheng, Y, Garavito, R.M, Ferguson-Miller, S. | | Deposit date: | 2014-08-13 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of translocator protein (TSPO) and mutant mimic of a human polymorphism.
Science, 347, 2015
|
|
4UHZ
 
 | | Crystal structure of the human RGMB-BMP2 complex, crystal form 1 | | Descriptor: | BONE MORPHOGENETIC PROTEIN 2, RGM DOMAIN FAMILY MEMBER B, SULFATE ION | | Authors: | Healey, E.G, Bishop, B, Elegheert, J, Bell, C.H, Padilla-Parra, S, Siebold, C. | | Deposit date: | 2015-03-27 | | Release date: | 2015-05-06 | | Last modified: | 2015-06-17 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Repulsive Guidance Molecule is a Structural Bridge between Neogenin and Bone Morphogenetic Protein.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4UMN
 
 | | Structure of a stapled peptide antagonist bound to Nutlin-resistant Mdm2. | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, M06 | | Authors: | Chee, S, Wongsantichon, J, Quah, S, Robinson, R.C, Verma, C, Lane, D.P, Brown, C.J, Ghadessy, F.J. | | Deposit date: | 2014-05-20 | | Release date: | 2014-05-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure of a stapled peptide antagonist bound to nutlin-resistant Mdm2.
PLoS ONE, 9, 2014
|
|
4UHY
 
 | | Crystal structure of the human RGMA-BMP2 complex | | Descriptor: | BONE MORPHOGENETIC PROTEIN 2, REPULSIVE GUIDANCE MOLECULE A | | Authors: | Healey, E.G, Bishop, B, Elegheert, J, Bell, C.H, Padilla-Parra, S, Siebold, C. | | Deposit date: | 2015-03-27 | | Release date: | 2015-05-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Repulsive Guidance Molecule is a Structural Bridge between Neogenin and Bone Morphogenetic Protein.
Nat.Struct.Mol.Biol., 22, 2015
|
|
3CEK
 
 | | Crystal structure of human dual specificity protein kinase (TTK) | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, Dual specificity protein kinase TTK | | Authors: | Filippakopoulos, P, Soundararajan, M, Keates, T, Elkins, J.M, King, O, Fedorov, O, Picaud, S.S, Pike, A.C.W, Roos, A, Pilka, E, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-02-29 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Small-molecule kinase inhibitors provide insight into Mps1 cell cycle function.
Nat.Chem.Biol., 6, 2010
|
|
7OT5
 
 | | CspA-70 cotranslational folding intermediate 1 | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Agirrezabala, X, Samatova, E, Macher, M, Liutkute, M, Gil-Carton, D, Novacek, J, Valle, M, Rodnina, M.V. | | Deposit date: | 2021-06-09 | | Release date: | 2022-01-19 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | A switch from alpha-helical to beta-strand conformation during co-translational protein folding.
Embo J., 41, 2022
|
|
4UOV
 
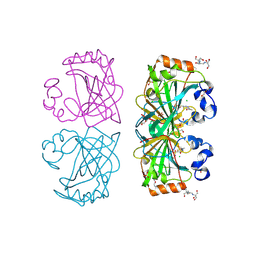 | | The structure of a tetrameric alpha-carbonic anhydrase from Thermovibrio ammonificans reveals a core formed around intermolecular disulfides, which contribute to its thermostability. | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, ... | | Authors: | James, P, Isupov, M, Sayer, C, Berg, S, Lioliou, M, Kotlar, H, Littlechild, J. | | Deposit date: | 2014-06-10 | | Release date: | 2014-10-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Structure of a Tetrameric [Alpha]-Carbonic Anhydrase from Thermovibrio Ammonificans Reveals a Core Formed Around Intermolecular Disulfides that Contribute to its Thermostability
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7OIF
 
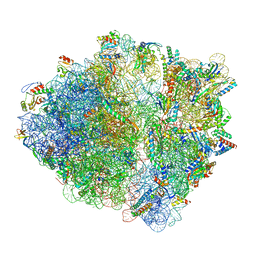 | | CspA-27 cotranslational folding intermediate 2 | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Agirrezabala, X, Samatova, E, Macher, M, Liutkute, M, Gil-Carton, D, Novacek, J, Valle, M, Rodnina, M.V. | | Deposit date: | 2021-05-11 | | Release date: | 2022-01-19 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A switch from alpha-helical to beta-strand conformation during co-translational protein folding.
Embo J., 41, 2022
|
|
7OIG
 
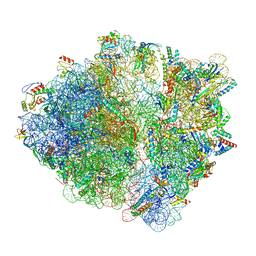 | | CspA-27 cotranslational folding intermediate 3 | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Agirrezabala, X, Samatova, E, Macher, M, Liutkute, M, Gil-Carton, D, Novacek, J, Valle, M, Rodnina, M.V. | | Deposit date: | 2021-05-11 | | Release date: | 2022-01-19 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A switch from alpha-helical to beta-strand conformation during co-translational protein folding.
Embo J., 41, 2022
|
|
7OII
 
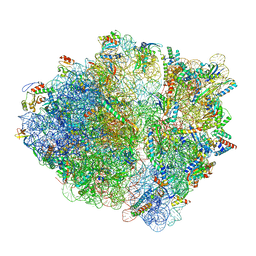 | | CspA-70 cotranslational folding intermediate 2 | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Agirrezabala, X, Samatova, E, Macher, M, Liutkute, M, Gil-Carton, D, Novacek, J, Valle, M, Rodnina, M.V. | | Deposit date: | 2021-05-11 | | Release date: | 2022-01-19 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A switch from alpha-helical to beta-strand conformation during co-translational protein folding.
Embo J., 41, 2022
|
|
4UI2
 
 | | Crystal structure of the ternary RGMB-BMP2-NEO1 complex | | Descriptor: | ACETATE ION, BONE MORPHOGENETIC PROTEIN 2, BMP2, ... | | Authors: | Healey, E.G, Bishop, B, Elegheert, J, Bell, C.H, Padilla-Parra, S, Siebold, C. | | Deposit date: | 2015-03-27 | | Release date: | 2015-05-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Repulsive Guidance Molecule is a Structural Bridge between Neogenin and Bone Morphogenetic Protein.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4UN8
 
 | | THE CRYSTAL STRUCTURE OF I-DMOI IN COMPLEX WITH ITS TARGET DNA AT 1H INCUBATION IN 5MM MN (STATE 2) | | Descriptor: | 25MER, HOMING ENDONUCLEASE I-DMOI, MANGANESE (II) ION | | Authors: | Molina, R, Stella, S, Redondo, P, Gomez, H, Marcaida, M.J, Orozco, M, Prieto, J, Montoya, G. | | Deposit date: | 2014-05-26 | | Release date: | 2014-12-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Visualizing Phosphodiester-Bond Hydrolysis by an Endonuclease.
Nat.Struct.Mol.Biol., 22, 2015
|
|
