7QCE
 
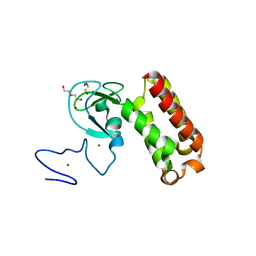 | | Crystal structure of an atypical PHD finger of VIN3 | | Descriptor: | DI(HYDROXYETHYL)ETHER, VIN3 (Protein VERNALIZATION INSENSITIVE 3), ZINC ION | | Authors: | Franco-Echevarria, E, Fiedler, M, Dean, C, Bienz, M. | | Deposit date: | 2021-11-23 | | Release date: | 2022-11-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Plant vernalization proteins contain unusual PHD superdomains without histone H3 binding activity.
J.Biol.Chem., 298, 2022
|
|
4UPU
 
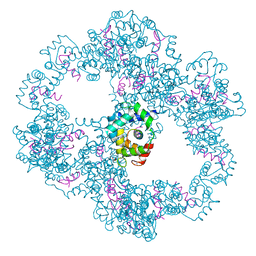 | | Crystal structure of IP3 3-K calmodulin binding region in complex with Calmodulin | | Descriptor: | CALCIUM ION, CALMODULIN, GLYCEROL, ... | | Authors: | Franco-Echevarria, E, Banos-Sanz, J.I, Monterroso, B, Round, A, Sanz-Aparicio, J, Gonzalez, B. | | Deposit date: | 2014-06-18 | | Release date: | 2014-08-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | A New Calmodulin Binding Motif for Inositol 1,4,5-Trisphosphate 3-Kinase Regulation.
Biochem.J., 463, 2014
|
|
5MW8
 
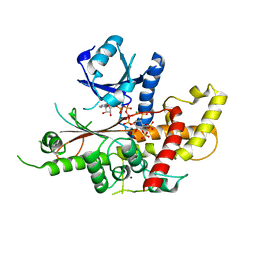 | | INOSITOL 1,3,4,5,6-PENTAKISPHOSPHATE 2-KINASE FROM M. MUSCULUS IN COMPLEX WITH ATP and IP5 | | Descriptor: | ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, Inositol-pentakisphosphate 2-kinase, ... | | Authors: | Franco-Echevarria, E, Sanz-Aparicio, J, Gonzalez, B. | | Deposit date: | 2017-01-18 | | Release date: | 2017-05-10 | | Last modified: | 2017-07-05 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of mammalian inositol 1,3,4,5,6-pentakisphosphate 2-kinase reveals a new zinc-binding site and key features for protein function.
J. Biol. Chem., 292, 2017
|
|
5MWL
 
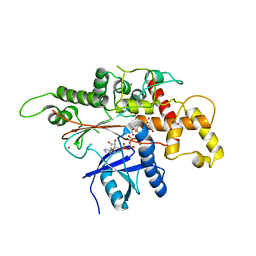 | | INOSITOL 1,3,4,5,6-PENTAKISPHOSPHATE 2-KINASE FROM M. MUSCULUS IN COMPLEX WITH ATP and IP5 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Inositol-pentakisphosphate 2-kinase, MAGNESIUM ION, ... | | Authors: | Franco-Echevarria, E, Sanz-Aparicio, J, Gonzalez, B. | | Deposit date: | 2017-01-18 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The crystal structure of mammalian inositol 1,3,4,5,6-pentakisphosphate 2-kinase reveals a new zinc-binding site and key features for protein function.
J. Biol. Chem., 292, 2017
|
|
5MWM
 
 | | INOSITOL 1,3,4,5,6-PENTAKISPHOSPHATE 2-KINASE FROM M. MUSCULUS IN COMPLEX WITH IP6 | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, Inositol-pentakisphosphate 2-kinase, ZINC ION | | Authors: | Franco-Echevarria, E, Sanz-Aparicio, J, Gonzalez, B. | | Deposit date: | 2017-01-18 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of mammalian inositol 1,3,4,5,6-pentakisphosphate 2-kinase reveals a new zinc-binding site and key features for protein function.
J. Biol. Chem., 292, 2017
|
|
5O20
 
 | | Structure of Nrd1 RNA binding domain in complex with RNA (UUAGUAAUCC) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Protein NRD1, RNA (5'-R(*UP*AP*GP*UP*AP*AP*UP*C)-3') | | Authors: | Franco-Echevarria, E, Perez-Canadillas, J.M, Gonzalez, B. | | Deposit date: | 2017-05-19 | | Release date: | 2017-08-02 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (3.53 Å) | | Cite: | The structure of transcription termination factor Nrd1 reveals an original mode for GUAA recognition.
Nucleic Acids Res., 45, 2017
|
|
5O1Y
 
 | | Structure of Nrd1 RNA binding domain in complex with RNA (GUAA) | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Protein NRD1, ... | | Authors: | Franco-Echevarria, E, Perez-Canadillas, J.M, Gonzalez, B. | | Deposit date: | 2017-05-19 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The structure of transcription termination factor Nrd1 reveals an original mode for GUAA recognition.
Nucleic Acids Res., 45, 2017
|
|
5O1X
 
 | | Structure of Nrd1 RNA binding domain | | Descriptor: | 1,2-ETHANEDIOL, Protein NRD1, THIOCYANATE ION | | Authors: | Franco-Echevarria, E, Perez-Canadillas, J.M, Gonzalez, B. | | Deposit date: | 2017-05-19 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure of transcription termination factor Nrd1 reveals an original mode for GUAA recognition.
Nucleic Acids Res., 45, 2017
|
|
5O1W
 
 | |
5O1Z
 
 | |
7O6W
 
 | | Crystal structure of (the) VEL1 VEL polymerising domain (I664D mutant) | | Descriptor: | PHOSPHATE ION, VIN3-like protein 2 | | Authors: | Fiedler, M, Franco-Echevarria, E, Dean, C, Bienz, M. | | Deposit date: | 2021-04-12 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-23 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Head-to-tail polymerization by VEL proteins underpins cold-induced Polycomb silencing in flowering control.
Cell Rep, 41, 2022
|
|
7O6V
 
 | |
7O6T
 
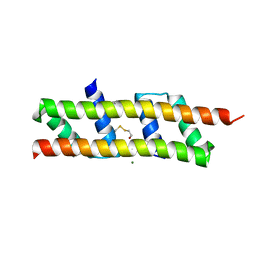 | | Crystal structure of the polymerising VEL domain of VIN3 (R556D I575D mutant) | | Descriptor: | MAGNESIUM ION, Protein VERNALIZATION INSENSITIVE 3 | | Authors: | Fiedler, M, Franco-Echevarria, E, Dean, C, Bienz, M. | | Deposit date: | 2021-04-12 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-23 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Head-to-tail polymerization by VEL proteins underpins cold-induced Polycomb silencing in flowering control.
Cell Rep, 41, 2022
|
|
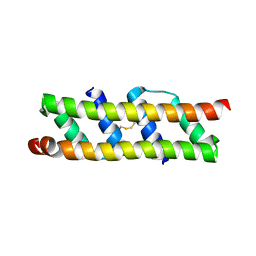
7OQV
 
 | |
7O6U
 
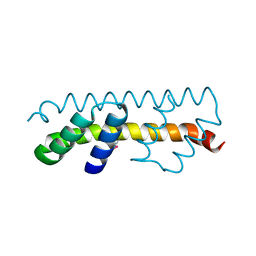 | |
