2VD7
 
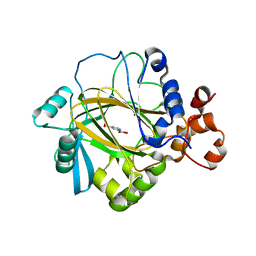 | | Crystal Structure of JMJD2A complexed with inhibitor Pyridine-2,4- dicarboxylic acid | | Descriptor: | JMJC DOMAIN-CONTAINING HISTONE DEMETHYLATION PROTEIN 3A, NICKEL (II) ION, PYRIDINE-2,4-DICARBOXYLIC ACID, ... | | Authors: | Ng, S.S, von Delft, F, Pilka, E.S, Kavanagh, K.L, McDonough, M.A, Savitsky, P, Arrowsmith, C.H, Weigelt, J, Edwards, A, Sundstrom, M, Schofield, C.J, Oppermann, U. | | Deposit date: | 2007-10-01 | | Release date: | 2007-11-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Inhibitor scaffolds for 2-oxoglutarate-dependent histone lysine demethylases.
J. Med. Chem., 51, 2008
|
|
5H56
 
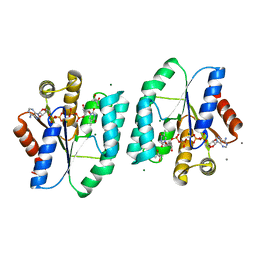 | | ADP and dTDP bound Crystal structure of thymidylate kinase (aq_969) from Aquifex Aeolicus VF5 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Biswas, A, Jeyakanthan, J, Sekar, K, Kuramitsu, S, Yokoyama, S. | | Deposit date: | 2016-11-04 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural studies of a hyperthermophilic thymidylate kinase enzyme reveal conformational substates along the reaction coordinate
FEBS J., 284, 2017
|
|
3UUE
 
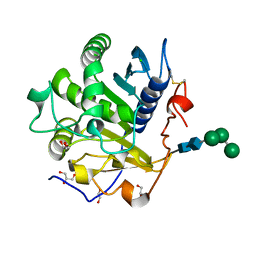 | | Crystal structure of mono- and diacylglycerol lipase from Malassezia globosa | | Descriptor: | CHLORIDE ION, GLYCEROL, LIP1, ... | | Authors: | Xu, T, Xu, J, Hou, S, Liu, J. | | Deposit date: | 2011-11-28 | | Release date: | 2012-04-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of a mono- and diacylglycerol lipase from Malassezia globosa reveals a novel lid conformation and insights into the substrate specificity.
J.Struct.Biol., 178, 2012
|
|
5GOO
 
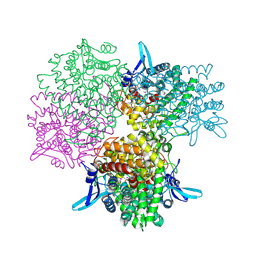 | | Crystal structure of alkaline invertase InvA from Anabaena sp. PCC 7120 complexed with fructose | | Descriptor: | Alkaline Invertase, GLYCEROL, beta-D-fructofuranose | | Authors: | Xie, J, Cai, K, Hu, H.X, Jiang, Y.L, Yang, F, Hu, P.F, Chen, Y, Zhou, C.Z. | | Deposit date: | 2016-07-28 | | Release date: | 2016-11-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural Analysis of the Catalytic Mechanism and Substrate Specificity of Anabaena Alkaline Invertase InvA Reveals a Novel Glucosidase.
J. Biol. Chem., 291, 2016
|
|
2WZL
 
 | | The Structure of the N-RNA Binding Domain of the Mokola virus Phosphoprotein | | Descriptor: | GLYCEROL, PHOSPHOPROTEIN | | Authors: | Assenberg, R, Delmas, O, Ren, J, Vidalain, P, Verma, A, Larrous, F, Graham, S, Tangy, F, Grimes, J, Bourhy, H. | | Deposit date: | 2009-11-30 | | Release date: | 2009-12-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structure of the N-RNA Binding Domain of the Mokola Virus Phosphoprotein
J.Virol., 84, 2010
|
|
4OPE
 
 | | Streptomcyes albus JA3453 oxazolomycin ketosynthase domain OzmH KS7 | | Descriptor: | NITRATE ION, NRPS/PKS | | Authors: | Osipiuk, J, Mack, J, Endres, M, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-02-05 | | Release date: | 2014-02-19 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural and evolutionary relationships of "AT-less" type I polyketide synthase ketosynthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4A61
 
 | | ParM from plasmid R1 in complex with AMPPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PLASMID SEGREGATION PROTEIN PARM | | Authors: | van den Ent, F, Moller-Jensen, J, Gayathri, P, Lowe, J. | | Deposit date: | 2011-10-31 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Bipolar Spindle of Antiparallel Parm Filaments Drives Bacterial Plasmid Segregation.
Science, 338, 2012
|
|
3TI5
 
 | | Crystal structure of 2009 pandemic H1N1 neuraminidase complexed with Zanamivir | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CALCIUM ION, ... | | Authors: | Vavricka, C.J, Li, Q, Wu, Y, Qi, J, Wang, M, Liu, Y, Gao, F, Liu, J, Feng, E, He, J, Wang, J, Liu, H, Jiang, H, Gao, G.F. | | Deposit date: | 2011-08-20 | | Release date: | 2011-11-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional analysis of laninamivir and its octanoate prodrug reveals group specific mechanisms for influenza NA inhibition
Plos Pathog., 7, 2011
|
|
3TI6
 
 | | Crystal structure of 2009 pandemic H1N1 neuraminidase complexed with oseltamivir | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Vavricka, C.J, Li, Q, Wu, Y, Qi, J, Wang, M, Liu, Y, Gao, F, Liu, J, Feng, E, He, J, Wang, J, Liu, H, Jiang, H, Gao, G.F. | | Deposit date: | 2011-08-20 | | Release date: | 2011-11-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural and functional analysis of laninamivir and its octanoate prodrug reveals group specific mechanisms for influenza NA inhibition
Plos Pathog., 7, 2011
|
|
3OF1
 
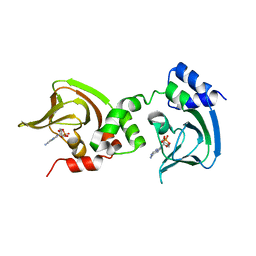 | | Crystal Structure of Bcy1, the Yeast Regulatory Subunit of PKA | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, cAMP-dependent protein kinase regulatory subunit | | Authors: | Rinaldi, J, Wu, J, Yang, J, Ralston, C.Y, Sankaran, B, Moreno, S, Taylor, S.S. | | Deposit date: | 2010-08-13 | | Release date: | 2010-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure of Yeast Regulatory Subunit: A Glimpse into the Evolution of PKA Signaling.
Structure, 18, 2010
|
|
3L3J
 
 | | Crystal structure of HLA-B*4402 in complex with the F3A/R5A double mutant of a self-peptide derived from DPA*0201 | | Descriptor: | ACETATE ION, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Theodossis, A, Ely, L.K, Rossjohn, J. | | Deposit date: | 2009-12-17 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Constraints within major histocompatibility complex class I restricted peptides: presentation and consequences for T-cell recognition
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3L3U
 
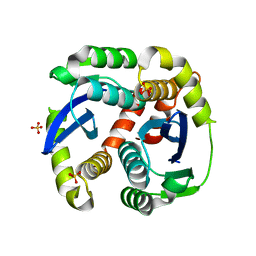 | | Crystal structure of the HIV-1 integrase core domain to 1.4A | | Descriptor: | POL polyprotein, SULFATE ION | | Authors: | Wielens, J, Chalmers, D.K, Scanlon, M.J, Parker, M.W. | | Deposit date: | 2009-12-17 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the HIV-1 integrase core domain in complex with sucrose reveals details of an allosteric inhibitory binding site.
Febs Lett., 584, 2010
|
|
5GYQ
 
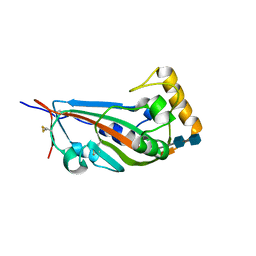 | |
4AHV
 
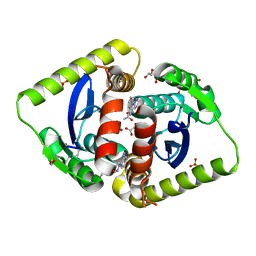 | | Parallel screening of a low molecular weight compound library: do differences in methodology affect hit identification | | Descriptor: | 1,2-ETHANEDIOL, 1-[2-(1H-pyrazol-1-yl)phenyl]methanamine, ACETIC ACID, ... | | Authors: | Wielens, J, Heady, S.J, Rhodes, D.I, Mulder, R.J, Dolezal, O, Deadman, J.J, Newman, J, Chalmers, D.K, Parker, M.W, Peat, T.S, Scanlon, M.J. | | Deposit date: | 2012-02-07 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Parallel Screening of Low Molecular Weight Fragment Libraries: Do Differences in Methodology Affect Hit Identification?
J.Biomol.Screen, 18, 2013
|
|
2Z6G
 
 | | Crystal Structure of a Full-Length Zebrafish Beta-Catenin | | Descriptor: | B-catenin | | Authors: | Xing, Y, Takemaru, K, Liu, J, Zheng, J, Moon, R, Xu, W. | | Deposit date: | 2007-08-01 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal Structure of a Full-Length beta-Catenin
Structure, 16, 2008
|
|
3F0W
 
 | | Human NUMB-like protein, phosphotyrosine interaction domain | | Descriptor: | CHLORIDE ION, Numb-like protein, SULFATE ION | | Authors: | Lehtio, L, Moche, M, Andersson, J, Arrowsmith, C.H, Berglund, H, Bountra, C, D Busam, R, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kotenyova, T, Nilsson, M.E, Nyman, T, Persson, C, Sagemark, J, Schueler, H, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Wikstrom, M, Wisniewska, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-10-27 | | Release date: | 2008-11-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Human NUMB-like protein, phosphotyrosine interaction domain
To be Published
|
|
4S38
 
 | | IspG in complex MEcPP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2C-METHYL-D-ERYTHRITOL 2,4-CYCLODIPHOSPHATE, 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase, ... | | Authors: | Quitterer, F, Frank, A, Wang, K, Guodong, R, O'Dowd, B, Li, J, Guerra, F, Abdel-Azeim, S, Bacher, A, Eppinger, J, Oldfield, E, Groll, M. | | Deposit date: | 2015-01-26 | | Release date: | 2015-04-29 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Atomic-Resolution Structures of Discrete Stages on the Reaction Coordinate of the [Fe4S4] Enzyme IspG (GcpE).
J.Mol.Biol., 427, 2015
|
|
2XB2
 
 | | Crystal structure of the core Mago-Y14-eIF4AIII-Barentsz-UPF3b assembly shows how the EJC is bridged to the NMD machinery | | Descriptor: | EUKARYOTIC INITIATION FACTOR 4A-III, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Buchwald, G, Ebert, J, Basquin, C, Sauliere, J, Jayachandran, U, Bono, F, Le Hir, H, Conti, E. | | Deposit date: | 2010-04-03 | | Release date: | 2010-05-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Insights Into the Recruitment of the Nmd Machinery from the Crystal Structure of a Core Ejc-Upf3B Complex.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4S3D
 
 | | IspG in complex with PPi | | Descriptor: | 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase, DIPHOSPHATE, FE3-S4 CLUSTER | | Authors: | Quitterer, F, Frank, A, Wang, K, Rao, G, O'Dowd, B, Li, J, Guerra, F, Abdel-Azeim, S, Bacher, A, Eppinger, J, Oldfield, E, Groll, M. | | Deposit date: | 2015-01-26 | | Release date: | 2015-04-29 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Atomic-Resolution Structures of Discrete Stages on the Reaction Coordinate of the [Fe4S4] Enzyme IspG (GcpE).
J.Mol.Biol., 427, 2015
|
|
4RK0
 
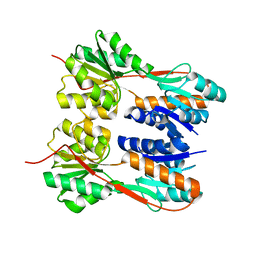 | | Crystal structure of LacI family transcriptional regulator from Enterococcus faecalis V583, Target EFI-512923, with bound ribose | | Descriptor: | LacI family sugar-binding transcriptional regulator, alpha-D-ribofuranose | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Siedel, R.D, Hillerich, B, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-10-11 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of LacI Transcriptional Regulator Ef_2962 from Enterococcus faecalis V583, Target EFI-512923
To be Published
|
|
3L3D
 
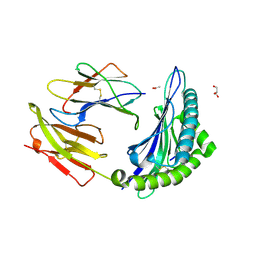 | | Crystal structure of HLA-B*4402 in complex with the F3A mutant of a self-peptide derived from DPA*0201 | | Descriptor: | ACETATE ION, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Theodossis, A, Ely, L.K, Rossjohn, J. | | Deposit date: | 2009-12-16 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Constraints within major histocompatibility complex class I restricted peptides: presentation and consequences for T-cell recognition
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3F8Z
 
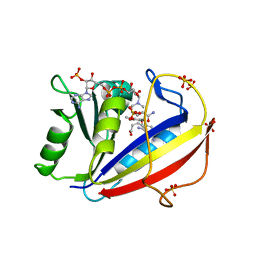 | | Human Dihydrofolate Reductase Structural Data with Active Site Mutant Enzyme Complexes | | Descriptor: | 2,4-DIAMINO-5-[2-METHOXY-5-(4-CARBOXYBUTYLOXY)BENZYL]PYRIMIDINE, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Cody, V, Pace, J, Makin, J, Piraino, J, Queener, S.F, Rosowsky, A. | | Deposit date: | 2008-11-13 | | Release date: | 2009-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Correlations of Inhibitor Kinetics for Pneumocystis jirovecii and Human Dihydrofolate Reductase with Structural Data for Human Active Site Mutant Enzyme Complexes.
Biochemistry, 48, 2009
|
|
5C5Y
 
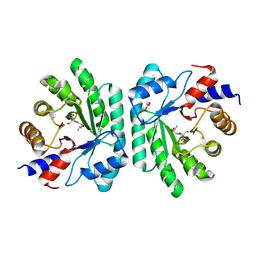 | |
3TIB
 
 | | Crystal structure of 1957 pandemic H2N2 neuraminidase complexed with laninamivir octanoate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-acetamido-2,6-anhydro-4-carbamimidamido-3,4,5-trideoxy-7-O-methyl-9-O-octanoyl-D-glycero-D-galacto-non-2-enonic acid, CALCIUM ION, ... | | Authors: | Vavricka, C.J, Li, Q, Wu, Y, Qi, J, Wang, M, Liu, Y, Gao, F, Liu, J, Feng, E, He, J, Wang, J, Liu, H, Jiang, H, Gao, G.F. | | Deposit date: | 2011-08-20 | | Release date: | 2011-11-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structural and functional analysis of laninamivir and its octanoate prodrug reveals group specific mechanisms for influenza NA inhibition
Plos Pathog., 7, 2011
|
|
3L3K
 
 | | Crystal structure of HLA-B*4402 in complex with the R5A/F7A double mutant of a self-peptide derived from DPA*0201 | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Theodossis, A, Ely, L.K, Rossjohn, J. | | Deposit date: | 2009-12-17 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Constraints within major histocompatibility complex class I restricted peptides: presentation and consequences for T-cell recognition
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
