5F1R
 
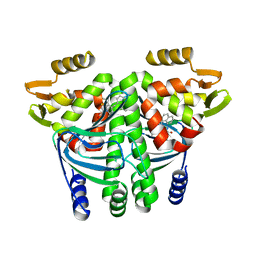 | | The Transcriptional Regulator PrfA from Listeria Monocytogenes in complex with a ring-fused 2-pyridone (C10) | | Descriptor: | (3~{R})-8-cyclopropyl-7-(naphthalen-1-ylmethyl)-5-oxidanylidene-2,3-dihydro-[1,3]thiazolo[3,2-a]pyridine-3-carboxylic acid, Listeriolysin regulatory protein | | Authors: | Begum, A, Grundstrom, C, Good, J.A.D, Andersson, C, ALmqvist, F, Johansson, J, Sauer, U.H, Sauer-Eriksson, A.E. | | Deposit date: | 2015-11-30 | | Release date: | 2016-03-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Attenuating Listeria monocytogenes Virulence by Targeting the Regulatory Protein PrfA.
Cell Chem Biol, 23, 2016
|
|
8EO6
 
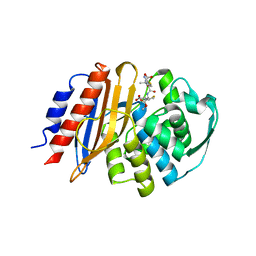 | | Crystal structure of metagenomic class A beta-lactamase precursor LRA-5 in complex with ceftazidime at 2.35 Angstrom resolution | | Descriptor: | ACYLATED CEFTAZIDIME, LRA-5 | | Authors: | Power, P, D'Amico Gonzalez, G, Centron, D, Gutkind, G, Handelsman, J, Klinke, S. | | Deposit date: | 2022-10-02 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Playing beta-Lactamase Evolution: Metagenomic Class A beta-Lactamase LRA-5 is an Inactive Enzyme Capable of Rendering an Active beta-Lactamase by Introduction of Y69Q and V166E Substitutions
to be published
|
|
6IFK
 
 | | Cryo-EM structure of type III-A Csm-CTR1 complex, AMPPNP bound | | Descriptor: | CTR1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | You, L, Ma, J, Wang, J, Zhang, X, Wang, Y. | | Deposit date: | 2018-09-20 | | Release date: | 2018-12-12 | | Last modified: | 2019-01-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure Studies of the CRISPR-Csm Complex Reveal Mechanism of Co-transcriptional Interference
Cell, 176, 2019
|
|
3RGZ
 
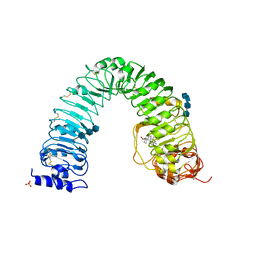 | | Structural insight into brassinosteroid perception by BRI1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chai, J, Han, Z, She, J, Wang, J, Cheng, W. | | Deposit date: | 2011-04-11 | | Release date: | 2011-06-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.281 Å) | | Cite: | Structural insight into brassinosteroid perception by BRI1.
Nature, 474, 2011
|
|
5LCH
 
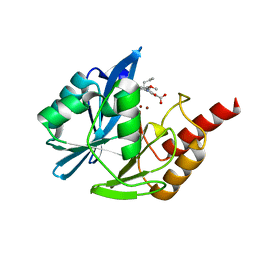 | | VIM-2 metallo-beta-lactamase in complex with (S)-1-allyl-2-(3-methoxyphenyl)-3-oxoisoindoline-4-carboxylic acid (compound 42) | | Descriptor: | (1~{S})-2-(3-methoxyphenyl)-3-oxidanylidene-1-prop-2-enyl-1~{H}-isoindole-4-carboxylic acid, Metallo-beta-lactamase VIM-2, ZINC ION | | Authors: | Li, G.-B, Brem, J, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2016-06-21 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | NMR-filtered virtual screening leads to non-metal chelating metallo-beta-lactamase inhibitors.
Chem Sci, 8, 2017
|
|
4Q9W
 
 | |
5LHK
 
 | | Bottromycin maturation enzyme BotP in complex with Mn | | Descriptor: | BICARBONATE ION, Leucine aminopeptidase 2, chloroplastic, ... | | Authors: | Koehnke, J, Adam, S. | | Deposit date: | 2016-07-12 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structure and Substrate Recognition of the Bottromycin Maturation Enzyme BotP.
Chembiochem, 17, 2016
|
|
4MD4
 
 | | Immune Receptor | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scally, S.W, Rossjohn, J. | | Deposit date: | 2013-08-22 | | Release date: | 2013-12-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A molecular basis for the association of the HLA-DRB1 locus, citrullination, and rheumatoid arthritis.
J.Exp.Med., 210, 2013
|
|
1TR1
 
 | | CRYSTAL STRUCTURE OF E96K MUTATED BETA-GLUCOSIDASE A FROM BACILLUS POLYMYXA, AN ENZYME WITH INCREASED THERMORESISTANCE | | Descriptor: | BETA-GLUCOSIDASE A, GLYCEROL | | Authors: | Sanz-Aparicio, J, Hermoso, J.A, Martinez-Ripoll, M, Gonzalez-Perez, B, Polaina, J. | | Deposit date: | 1998-03-12 | | Release date: | 1999-04-20 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of beta-glucosidase A from Bacillus polymyxa: insights into the catalytic activity in family 1 glycosyl hydrolases.
J.Mol.Biol., 275, 1998
|
|
5AMG
 
 | |
5LCA
 
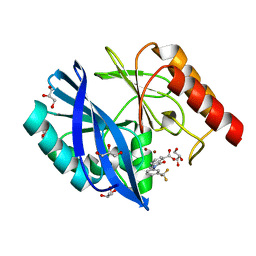 | | VIM-2 metallo-beta-lactamase in complex with 3-oxo-2-(3-(trifluoromethyl)phenyl)isoindoline-4-carboxylic acid (compound 17) | | Descriptor: | 3-oxidanylidene-2-[3-(trifluoromethyl)phenyl]-1~{H}-isoindole-4-carboxylic acid, GLYCEROL, Metallo-beta-lactamase VIM-2, ... | | Authors: | Li, G.-B, Brem, J, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2016-06-20 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | NMR-filtered virtual screening leads to non-metal chelating metallo-beta-lactamase inhibitors.
Chem Sci, 8, 2017
|
|
6I7B
 
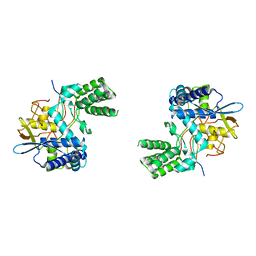 | | Influenza A nucleoprotein docked into 3D helical structure of the wild type ribonucleoprotein complex obtained using cryoEM. Conformation 3. | | Descriptor: | Nucleoprotein | | Authors: | Coloma, R, Arranz, R, de la Rosa-Trevin, J.M, Sorzano, C.O.S, Carlero, D, Ortin, J, Martin-Benito, J. | | Deposit date: | 2018-11-16 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Structural insights into influenza A virus ribonucleoproteins reveal a processive helical track as transcription mechanism.
Nat Microbiol, 5, 2020
|
|
4PVO
 
 | | Crystal Structure of VIM-2 metallo-beta-lactamase in complex with ML302 and ML302F | | Descriptor: | (2Z)-2-sulfanyl-3-(2,3,6-trichlorophenyl)prop-2-enoic acid, Beta-lactamase class B VIM-2, DIMETHYL SULFOXIDE, ... | | Authors: | Aik, W.S, Brem, J, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2014-03-18 | | Release date: | 2014-11-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Rhodanine hydrolysis leads to potent thioenolate mediated metallo-beta-lactamase inhibition.
Nat.Chem., 6, 2014
|
|
3VFM
 
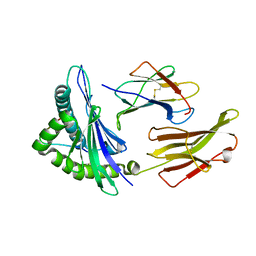 | | crystal structure of HLA B*3508 LPEP155A, HLA mutant Ala155 | | Descriptor: | Beta-2-microglobulin, LPEP peptide from EBV, LPEPLPQGQLTAY, ... | | Authors: | Liu, Y.C, Rossjohn, J, Gras, S. | | Deposit date: | 2012-01-10 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Energetic Basis Underpinning T-cell Receptor Recognition of a Super-bulged Peptide Bound to a Major Histocompatibility Complex Class I Molecule.
J.Biol.Chem., 287, 2012
|
|
4PX1
 
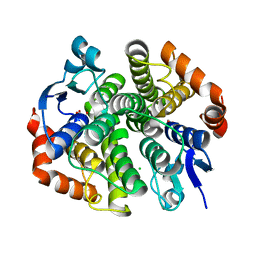 | | CRYSTAL STRUCTURE OF Maleylacetoacetate isomerase from Methylobacteriu extorquens AM1 WITH BOUND MALONATE (TARGET EFI-507068) | | Descriptor: | CHLORIDE ION, MALONIC ACID, Maleylacetoacetate isomerase (Glutathione S-transferase) | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-03-21 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of glutathione s-transferase zeta from methylobacterium extorquens (TARGET EFI-507068)
To be Published
|
|
8F1F
 
 | | Structure of K48-linked tri-ubiquitin in complex with cyclic peptide | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, Non-proteinogenic cyclic peptide (inhibitor), ... | | Authors: | Lubkowski, J, Fushman, D, Lemma, B. | | Deposit date: | 2022-11-05 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mechanism of selective recognition of Lys48-linked polyubiquitin by macrocyclic peptide inhibitors of proteasomal degradation.
Nat Commun, 14, 2023
|
|
2RHD
 
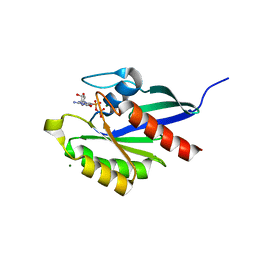 | | Crystal structure of Cryptosporidium parvum small GTPase RAB1A | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Small GTP binding protein rab1a | | Authors: | Dong, A, Xu, X, Lew, J, Lin, Y.H, Khuu, C, Sun, X, Qiu, W, Kozieradzki, I, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Sukumar, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-10-09 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal structure of Cryptosporidium parvum small GTPase RAB1A.
To be Published
|
|
3VRI
 
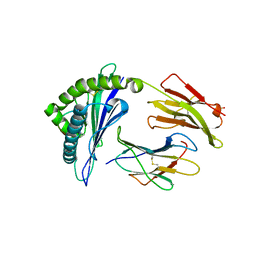 | | HLA-B*57:01-RVAQLENVYI in complex with abacavir | | Descriptor: | 10-mer peptide, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Vivian, J.P, Illing, P.T, McCluskey, J, Rossjohn, J. | | Deposit date: | 2012-04-11 | | Release date: | 2012-05-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Immune self-reactivity triggered by drug-modified HLA-peptide repertoire
Nature, 486, 2012
|
|
6IG0
 
 | | Type III-A Csm complex, Cryo-EM structure of Csm-CTR1, ATP bound | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CTR1, MAGNESIUM ION, ... | | Authors: | You, L, Ma, J, Wang, J, Zhang, X, Wang, Y. | | Deposit date: | 2018-09-21 | | Release date: | 2018-12-12 | | Last modified: | 2019-01-23 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Structure Studies of the CRISPR-Csm Complex Reveal Mechanism of Co-transcriptional Interference
Cell, 176, 2019
|
|
1IZI
 
 | | Inhibitor of HIV protease with unusual binding mode potently inhibiting multi-resistant protease mutants | | Descriptor: | CHLORIDE ION, proteinase, {(1S)-1-BENZYL-4-[3-CARBAMOYL-1-(1-CARBAMOYL-2-PHENYL-ETHYLCARBAMOYL)-(S)-PROPYLCARBAMOYL]-2-OXO-5-PHENYL-PENTYL}-CARBAMIC ACID TERT-BUTYL ESTER | | Authors: | Weber, J, Mesters, J.R, Lepsik, M, Prejdova, J, Svec, M, Sponarova, J, Mlcochova, P, Skalicka, K, Strisovsky, K, Uhlikova, T, Soucek, M, Machala, L, Stankova, M, Vondrasek, J, Klimkait, T, Kraeusslich, H.-G, Hilgenfeld, R, Konvalinka, J. | | Deposit date: | 2002-10-02 | | Release date: | 2002-12-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Unusual Binding Mode of an HIV-1 Protease Inhibitor Explains its Potency against Multi-drug-resistant Virus Strains
J.MOL.BIOL., 324, 2002
|
|
5LM3
 
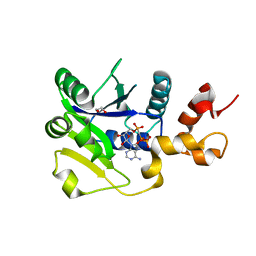 | | Plasmodium falciparum nicotinic acid mononucleotide adenylyltransferase complexed with APC | | Descriptor: | 1,2-ETHANEDIOL, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, Nicotinate-nucleotide adenylyltransferase | | Authors: | Bathke, J, Fritz-Wolf, K, Rahlfs, S, Brandstaether, C, Burkhardt, A, Becker, K. | | Deposit date: | 2016-07-29 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Functional Characterization of Plasmodium falciparum Nicotinic Acid Mononucleotide Adenylyltransferase.
J. Mol. Biol., 428, 2016
|
|
5AR7
 
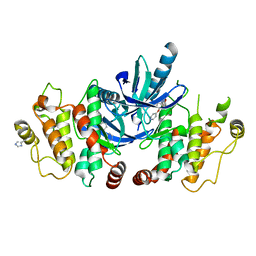 | | RIP2 Kinase Catalytic Domain (1 - 310) complex with Biaryl Urea | | Descriptor: | 1-(5-TERT-BUTYL-1,2-OXAZOL-3-YL)-3-(4-PYRIDIN-4-YLOXYPHENYL)UREA, RECEPTOR-INTERACTING SERINE/THREONINE-PROTEIN KINASE 2 | | Authors: | Charnley, A.K, Convery, M.A, Lakdawala Shah, A, Jones, E, Hardwicke, P, Bridges, A, Votta, B.J, Gough, P.J, Marquis, R.W, Bertin, J, Casillas, L. | | Deposit date: | 2015-09-24 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystal Structures of Human Rip2 Kinase Catalytic Domain Complexed with ATP-Competitive Inhibitors: Foundations for Understanding Inhibitor Selectivity.
Bioorg.Med.Chem., 23, 2015
|
|
4BV7
 
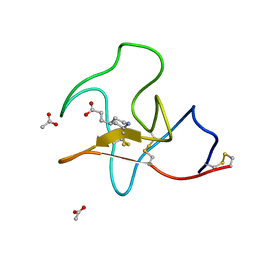 | | Identification of small molecule inhibitors selective for apo(a) kringles KIV-7, KIV-10 and KV. | | Descriptor: | 3-(4-piperidyl)propanoic acid, ACETATE ION, APOLIPOPROTEIN(A) | | Authors: | Sandmark, J, Althage, M, Andersson, G.M.K, Antonsson, T, Blaho, S, Bodin, C, Bostrom, J, Chen, Y, Dahlen, A, Eriksson, P.O, Evertsson, E, Fex, T, Fjellstrom, O, Gustafsson, D, Hallberg, C, Hicks, R, Jarkvist, E, Johansson, C, Kalies, I, Kang, D, Svalstedt Karlsson, B, Kartberg, F, Legnehed, A, Lindqvist, A.M, Martinsson, S.A, Moberg, A, Petersson, A.U, Ridderstrom, M, Thelin, A, Tigerstrom, A, Vinblad, J, Xu, B, Knecht, W. | | Deposit date: | 2013-06-25 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Small Molecules Used to Decipher the Pathophysiological Roles of the Kringle Domains Kiv-7, - 10 and Kv of Apolipoprotein(A)
To be Published
|
|
2Z6H
 
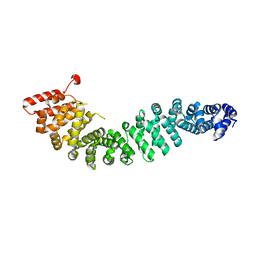 | | Crystal Structure of Beta-Catenin Armadillo Repeat Region and Its C-Terminal domain | | Descriptor: | Catenin beta-1 | | Authors: | Xing, Y, Takemaru, K, Liu, J, Zheng, J, Moon, R, Xu, W. | | Deposit date: | 2007-08-01 | | Release date: | 2008-02-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of a Full-Length beta-Catenin
Structure, 16, 2008
|
|
2RT8
 
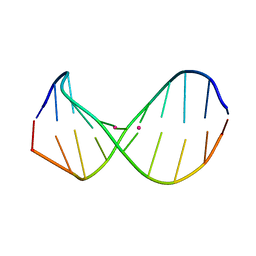 | | Structure of metallo-dna in solution | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*TP*TP*GP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*CP*TP*TP*CP*GP*CP*G)-3'), MERCURY (II) ION | | Authors: | Yamaguchi, H, Sebera, J, Kondo, J, Oda, S, Komuro, T, Kawamura, T, Dairaku, T, Kondo, Y, Okamoto, I, Ono, A, Burda, J.V, Kojima, C, Sychrovsky, V, Tanaka, Y. | | Deposit date: | 2013-06-18 | | Release date: | 2014-03-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The structure of metallo-DNA with consecutive thymine-HgII-thymine base pairs explains positive entropy for the metallo base pair formation.
Nucleic Acids Res., 42, 2014
|
|
