6A5W
 
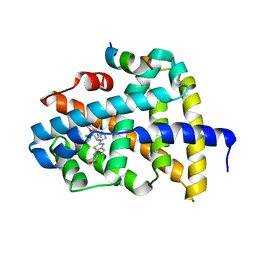 | | FXR-LBD with HNC143 and SRC1 | | Descriptor: | 2-[2-[[3-[2,6-bis(chloranyl)phenyl]-5-cyclopropyl-1,2-oxazol-4-yl]methoxy]-6-azaspiro[3.4]octan-6-yl]-1,3-benzothiazole-6-carboxylic acid, Bile acid receptor, Nuclear receptor coactivator 1 | | Authors: | Wang, N, Liu, J. | | Deposit date: | 2018-06-25 | | Release date: | 2018-10-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Ligand binding and heterodimerization with retinoid X receptor alpha (RXR alpha ) induce farnesoid X receptor (FXR) conformational changes affecting coactivator binding
J. Biol. Chem., 293, 2018
|
|
8T9L
 
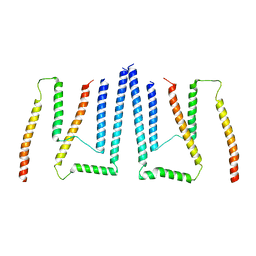 | | Pom34-Pom152 membrane attachment site yeast NPC | | Descriptor: | Nucleoporin POM152, Nucleoporin POM34 | | Authors: | Akey, C.W, Echeverria, I, Ouch, C, Fernandez-Martinez, J, Rout, M.P. | | Deposit date: | 2023-06-24 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Implications of a multiscale structure of the yeast nuclear pore complex.
Mol.Cell, 83, 2023
|
|
7Y72
 
 | | SARS-CoV-2 spike glycoprotein trimer complexed with Fab fragment of anti-RBD antibody E7 (focused refinement on Fab-RBD interface) | | Descriptor: | Fab E7 heavy chain, Fab E7 light chain, Spike glycoprotein | | Authors: | Chia, W.N, Tan, C.W, Tan, A.W.K, Young, B, Starr, T.N, Lopez, E, Fibriansah, G, Barr, J, Cheng, S, Yeoh, A.Y.Y, Yap, W.C, Lim, B.L, Ng, T.S, Sia, W.R, Zhu, F, Chen, S, Zhang, J, Greaney, A.J, Chen, M, Au, G.G, Paradkar, P, Peiris, M, Chung, A.W, Bloom, J.D, Lye, D, Lok, S.M, Wang, L.F. | | Deposit date: | 2022-06-21 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (4.03 Å) | | Cite: | Potent pan huACE2-dependent sarbecovirus neutralizing monoclonal antibodies isolated from a BNT162b2-vaccinated SARS survivor.
Sci Adv, 9, 2023
|
|
8TJ5
 
 | | Inner spoke ring of the yeast NPC | | Descriptor: | Nucleoporin 59, Nucleoporin NIC96, Nucleoporin NSP1, ... | | Authors: | Akey, C.W, Echeverria, I, Ouch, C, Fernandez-Martinez, J, Rout, M.P. | | Deposit date: | 2023-07-20 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Implications of a multiscale structure of the yeast nuclear pore complex.
Mol.Cell, 83, 2023
|
|
8TIE
 
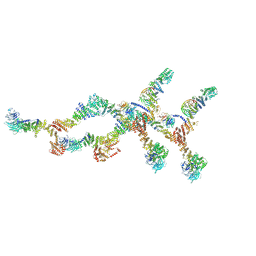 | | Double nuclear outer ring of Nup84-complexes from the yeast NPC | | Descriptor: | NUP133 isoform 1, NUP145 isoform 1, Nucleoporin NUP120, ... | | Authors: | Akey, C.W, Echeverria, I, Ouch, C, Fernandez-Martinez, J, Rout, M.P. | | Deposit date: | 2023-07-19 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (8.1 Å) | | Cite: | Implications of a multiscale structure of the yeast nuclear pore complex.
Mol.Cell, 83, 2023
|
|
6U4J
 
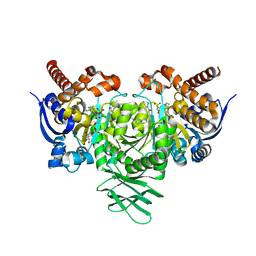 | | Crystal structure of IDH1 R132H mutant in complex with FT-2102 | | Descriptor: | 5-{[(1S)-1-(6-chloro-2-oxo-1,2-dihydroquinolin-3-yl)ethyl]amino}-1-methyl-6-oxo-1,6-dihydropyridine-2-carbonitrile, CHLORIDE ION, CITRATE ANION, ... | | Authors: | Toms, A.V, Lin, J. | | Deposit date: | 2019-08-25 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structure-Based Design and Identification of FT-2102 (Olutasidenib), a Potent Mutant-Selective IDH1 Inhibitor.
J.Med.Chem., 63, 2020
|
|
7Y71
 
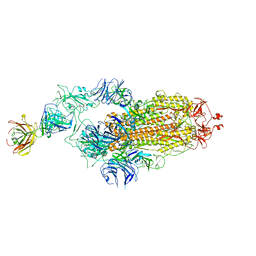 | | SARS-CoV-2 spike glycoprotein trimer complexed with Fab fragment of anti-RBD antibody E7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab E7 heavy chain, ... | | Authors: | Chia, W.N, Tan, C.W, Tan, A.W.K, Young, B, Starr, T.N, Lopez, E, Fibriansah, G, Barr, J, Cheng, S, Yeoh, A.Y.Y, Yap, W.C, Lim, B.L, Ng, T.S, Sia, W.R, Zhu, F, Chen, S, Zhang, J, Greaney, A.J, Chen, M, Au, G.G, Paradkar, P, Peiris, M, Chung, A.W, Bloom, J.D, Lye, D, Lok, S.M, Wang, L.F. | | Deposit date: | 2022-06-21 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Potent pan huACE2-dependent sarbecovirus neutralizing monoclonal antibodies isolated from a BNT162b2-vaccinated SARS survivor.
Sci Adv, 9, 2023
|
|
5HC2
 
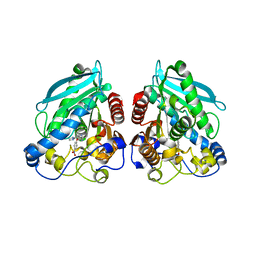 | |
2DH3
 
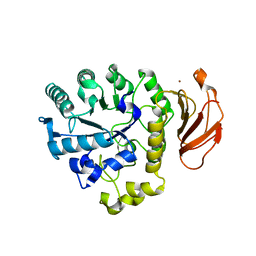 | | Crystal Structure of human ED-4F2hc | | Descriptor: | 4F2 cell-surface antigen heavy chain, ZINC ION | | Authors: | Fort, J, Fita, I, Palacin, M. | | Deposit date: | 2006-03-21 | | Release date: | 2007-03-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structure of human 4F2hc ectodomain provides a model for homodimerization and electrostatic interaction with plasma membrane.
J.Biol.Chem., 282, 2007
|
|
5HFK
 
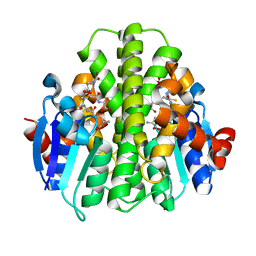 | | CRYSTAL STRUCTURE OF A GLUTATHIONE S-TRANSFERASE PROTEIN FROM ESCHERICHIA COLI OCh 157:H7 STR. SAKAI (ECs3186, TARGET EFI-507414) WITH BOUND GLUTATHIONE | | Descriptor: | Disulfide-bond oxidoreductase YfcG, GLUTATHIONE | | Authors: | Himmel, D.M, Toro, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Stead, M, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI), New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2016-01-07 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.551 Å) | | Cite: | CRYSTAL STRUCTURE OF A GLUTATHIONE S-TRANSFERASE PROTEIN FROM ESCHERICHIA COLI OCh 157:H7 STR. SAKAI (ECs3186, TARGET EFI-507414) WITH BOUND GLUTATHIONE
TO BE PUBLISHED
|
|
8T1R
 
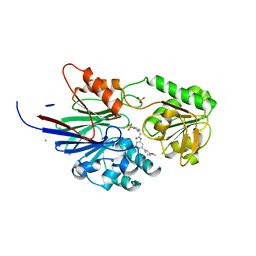 | | Crystal structure of human CPSF73 catalytic segment in complex with compound 2 | | Descriptor: | 3-[7,7-bis(oxidanyl)-8-oxa-7-boranuidabicyclo[4.3.0]nona-1,3,5-trien-5-yl]-~{N}-[3-(3-methoxyphenyl)phenyl]propanamide, CHLORIDE ION, Cleavage and polyadenylation specificity factor subunit 3, ... | | Authors: | Huang, J, Tong, L. | | Deposit date: | 2023-06-02 | | Release date: | 2023-11-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Anticancer benzoxaboroles block pre-mRNA processing by directly inhibiting CPSF3.
Cell Chem Biol, 31, 2024
|
|
8T1Q
 
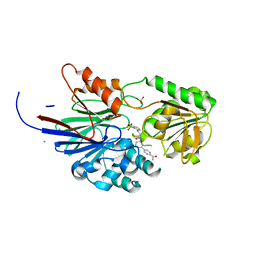 | | Crystal structure of human CPSF73 catalytic segment in complex with compound 1 | | Descriptor: | 3-[7,7-bis(oxidanyl)-8-oxa-7-boranuidabicyclo[4.3.0]nona-1,3,5-trien-5-yl]-~{N}-[3-(4-ethanoylphenyl)phenyl]propanamide, CHLORIDE ION, Cleavage and polyadenylation specificity factor subunit 3, ... | | Authors: | Huang, J, Tong, L. | | Deposit date: | 2023-06-02 | | Release date: | 2023-11-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Anticancer benzoxaboroles block pre-mRNA processing by directly inhibiting CPSF3.
Cell Chem Biol, 31, 2024
|
|
1A75
 
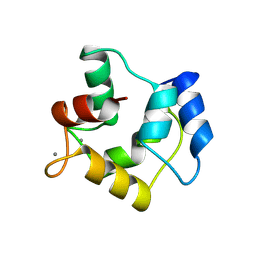 | | WHITING PARVALBUMIN | | Descriptor: | CALCIUM ION, PARVALBUMIN | | Authors: | Declercq, J.P, Baneres, J.L, Rambaud, J, Parello, J. | | Deposit date: | 1998-03-19 | | Release date: | 1998-06-17 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tertiary Structure of a Trp-Containing Parvalbumin from Whiting (Merlangius Merlangus). Description of the Hydrophobic Core
To be Published
|
|
5HC3
 
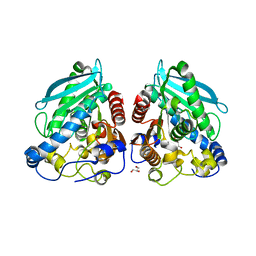 | | The structure of esterase Est22 | | Descriptor: | GLYCEROL, Lipolytic enzyme | | Authors: | Li, J, Huang, J. | | Deposit date: | 2016-01-04 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights of a hormone sensitive lipase homologue Est22.
Sci Rep, 6, 2016
|
|
5HSG
 
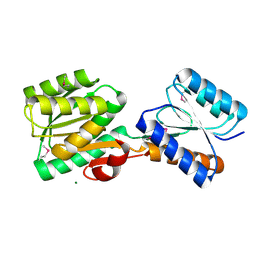 | | Crystal structure of an ABC transporter Solute Binding Protein from Klebsiella pneumoniae (KPN_01730, target EFI-511059), APO open structure | | Descriptor: | MAGNESIUM ION, Putative ABC transporter, nucleotide binding/ATPase protein | | Authors: | Roth, Y, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2016-01-25 | | Release date: | 2016-03-02 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of an ABC transporter Solute Binding Protein from Klebsiella pneumoniae (KPN_01730, target EFI-511059), APO open structure
To be published
|
|
6TWT
 
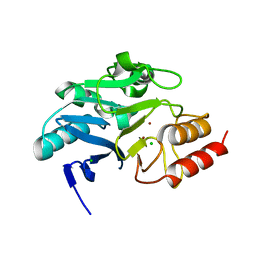 | | Crystal structure of N-terminally truncated NDM-1 metallo-beta-lactamase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Imiolczyk, B, Czyrko-Horczak, J, Brzezinski, K, Jaskolski, M. | | Deposit date: | 2020-01-13 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Flexible loops of New Delhi metallo-beta-lactamase modulate its activity towards different substrates.
Int.J.Biol.Macromol., 158, 2020
|
|
3VCL
 
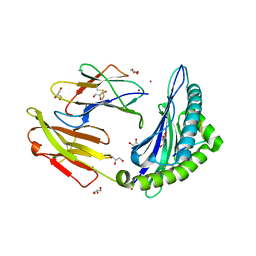 | |
6A5Y
 
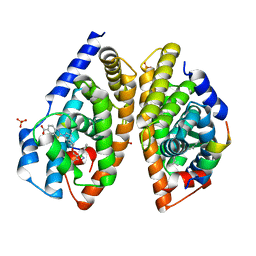 | | Crystal structure of human FXR/RXR-LBD heterodimer bound to HNC143 and 9cRA and SRC1 | | Descriptor: | (9cis)-retinoic acid, 2-[2-[[3-[2,6-bis(chloranyl)phenyl]-5-cyclopropyl-1,2-oxazol-4-yl]methoxy]-6-azaspiro[3.4]octan-6-yl]-1,3-benzothiazole-6-carboxylic acid, Bile acid receptor, ... | | Authors: | Wang, N, Liu, J. | | Deposit date: | 2018-06-25 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ligand binding and heterodimerization with retinoid X receptor alpha (RXR alpha ) induce farnesoid X receptor (FXR) conformational changes affecting coactivator binding
J. Biol. Chem., 293, 2018
|
|
5HKO
 
 | | Crystal structure of ABC transporter Solute Binding Protein MSMEG_3598 from Mycobacterium smegmatis str. MC2 155, target EFI-510969, in complex with L-sorbitol | | Descriptor: | ABC transporter, carbohydrate uptake transporter-2 (CUT2) family, periplasmic sugar-binding protein, ... | | Authors: | Roth, Y, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2016-01-14 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of ABC transporter Solute Binding Protein MSMEG_3598 from Mycobacterium smegmatis str. MC2 155, target EFI-510969, in complex with L-sorbitol
To be published
|
|
7ZEJ
 
 | | Crystal structure of the human MGC45594 gene product in complex with celecoxib. | | Descriptor: | 4-[5-(4-METHYLPHENYL)-3-(TRIFLUOROMETHYL)-1H-PYRAZOL-1-YL]BENZENESULFONAMIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Prostaglandin reductase 3 | | Authors: | Shafqat, N, Yue, W.W, Koekemoer, L, Niesen, F, Ugochukwu, E, Vollmar, M, Weigelt, J, Krojer, T, Pike, A, Chaikaud, A, Von Delft, F, Arrowsmith, C, Bountra, C, Edwards, A, Opperman, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-03-31 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Human prostaglandin/alkenal reductases: substrate specificities, inhibitor profiles, structural insights and subcellular localization suggest protective roles in inflammatory and oxidative stress conditions.
To Be Published
|
|
7ZCK
 
 | | Room temperature crystal structure of PhnD from Synechococcus MITS9220 in complex with phosphate | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, Phosphonate ABC type transporter/ substrate binding component | | Authors: | Mikolajek, H, Shah, B.S, Paulsen, I.T, Sandy, J, Sanchez-Weatherby, J. | | Deposit date: | 2022-03-28 | | Release date: | 2022-05-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Protein-to-structure pipeline for ambient-temperature in situ crystallography at VMXi.
Iucrj, 10, 2023
|
|
6A5X
 
 | | FXR-LBD with HNC180 and SRC1 | | Descriptor: | 2-[(1R,5S)-9-[[3-[2,6-bis(chloranyl)phenyl]-5-cyclopropyl-1,2-oxazol-4-yl]methoxy]-3-azabicyclo[3.3.1]nonan-3-yl]-1,3-benzothiazole-6-carboxylic acid, Bile acid receptor, Nuclear receptor coactivator 1, ... | | Authors: | Wang, N, Liu, J. | | Deposit date: | 2018-06-25 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Ligand binding and heterodimerization with retinoid X receptor alpha (RXR alpha ) induce farnesoid X receptor (FXR) conformational changes affecting coactivator binding
J. Biol. Chem., 293, 2018
|
|
8SQF
 
 | | OXA-48 bound to inhibitor CDD-2725 | | Descriptor: | (1M)-3'-(benzyloxy)-5-hydroxy[1,1'-biphenyl]-3,4'-dicarboxylic acid, BICARBONATE ION, Beta-lactamase | | Authors: | Park, S, Judge, A, Fan, J, Sankaran, B, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2023-05-04 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Exploiting the Carboxylate-Binding Pocket of beta-Lactamase Enzymes Using a Focused DNA-Encoded Chemical Library.
J.Med.Chem., 67, 2024
|
|
8SQG
 
 | | OXA-48 bound to inhibitor CDD-2801 | | Descriptor: | (1M)-3'-(benzyloxy)-5-[2-(methylamino)-2-oxoethoxy][1,1'-biphenyl]-3,4'-dicarboxylic acid, BICARBONATE ION, Beta-lactamase | | Authors: | Park, S, Judge, A, Fan, J, Sankaran, B, Palzkill, T. | | Deposit date: | 2023-05-04 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Exploiting the Carboxylate-Binding Pocket of beta-Lactamase Enzymes Using a Focused DNA-Encoded Chemical Library.
J.Med.Chem., 67, 2024
|
|
7ZBQ
 
 | | Structure of the ADP-ribosyltransferase TccC3HVR from Photorhabdus Luminescens | | Descriptor: | TccC3 | | Authors: | Lindemann, F, Belyy, A, Friedrich, D, Schmieder, P, Bardiaux, B, Roderer, D, Funk, J, Protze, J, Bieling, P, Oschkinat, H, Raunser, S. | | Deposit date: | 2022-03-24 | | Release date: | 2022-06-29 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Mechanism of threonine ADP-ribosylation of F-actin by a Tc toxin.
Nat Commun, 13, 2022
|
|
