8A4V
 
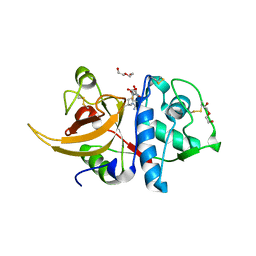 | | Crystal structure of human cathepsin L with covalently bound E-64 | | Descriptor: | Cathepsin L, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Falke, S, Lieske, J, Guenther, S, Reinke, P.Y.A, Ewert, W, Loboda, J, Karnicar, K, Usenik, A, Lindic, N, Sekirnik, A, Chapman, H.N, Hinrichs, W, Turk, D, Meents, A. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Elucidation and Antiviral Activity of Covalent Cathepsin L Inhibitors.
J.Med.Chem., 67, 2024
|
|
6GSZ
 
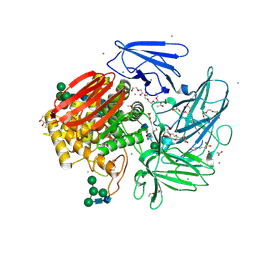 | | Crystal structure of native alfa-L-rhamnosidase from Aspergillus terreus | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, ... | | Authors: | Pachl, P, Rezacova, P, Skerlova, J. | | Deposit date: | 2018-06-15 | | Release date: | 2018-11-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Crystal structure of native alpha-L-rhamnosidase from Aspergillus terreus.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6GIR
 
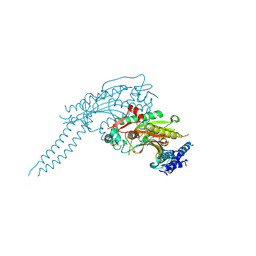 | | Arabidopsis thaliana cytosolic seryl-tRNA synthetase | | Descriptor: | Serine--tRNA ligase, cytoplasmic | | Authors: | Kekez, I, Kekez, M, Rokov-Plavec, J, Matkovic-Calogovic, D. | | Deposit date: | 2018-05-15 | | Release date: | 2019-01-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.343 Å) | | Cite: | Arabidopsis seryl-tRNA synthetase: the first crystal structure and novel protein interactor of plant aminoacyl-tRNA synthetase.
FEBS J., 286, 2019
|
|
8A4W
 
 | | Crystal structure of human cathepsin L with covalently bound Cathepsin L inhibitor IV | | Descriptor: | 1,2-ETHANEDIOL, Cathepsin L, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Falke, S, Lieske, J, Guenther, S, Reinke, P.Y.A, Ewert, W, Loboda, J, Karnicar, K, Usenik, A, Lindic, N, Sekirnik, A, Chapman, H.N, Hinrichs, W, Turk, D, Meents, A. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Elucidation and Antiviral Activity of Covalent Cathepsin L Inhibitors.
J.Med.Chem., 67, 2024
|
|
8A4X
 
 | | Crystal structure of human Cathepsin L with covalently bound Calpain inhibitor III | | Descriptor: | (phenylmethyl) N-[(2S)-3-methyl-1-oxidanylidene-1-[[(2S)-1-oxidanyl-3-phenyl-propan-2-yl]amino]butan-2-yl]carbamate, Cathepsin L, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Falke, S, Lieske, J, Guenther, S, Reinke, P.Y.A, Ewert, W, Loboda, J, Karnicar, K, Usenik, A, Lindic, N, Sekirnik, A, Chapman, H.N, Hinrichs, W, Turk, D, Meents, A. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Elucidation and Antiviral Activity of Covalent Cathepsin L Inhibitors.
J.Med.Chem., 67, 2024
|
|
8A4U
 
 | | Crystal structure of human cathepsin L with CAA0225 | | Descriptor: | (2S,3S)-N3-[2-(4-hydroxyphenyl)ethyl]-N2-[(2S)-1-oxidanylidene-3-phenyl-1-[(phenylmethyl)amino]propan-2-yl]oxirane-2,3-dicarboxamide, 1,2-ETHANEDIOL, Cathepsin L, ... | | Authors: | Falke, S, Lieske, J, Guenther, S, Reinke, P.Y.A, Ewert, W, Loboda, J, Karnicar, K, Usenik, A, Lindic, N, Sekirnik, A, Chapman, H.N, Hinrichs, W, Turk, D, Meents, A. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Elucidation and Antiviral Activity of Covalent Cathepsin L Inhibitors.
J.Med.Chem., 67, 2024
|
|
2CE6
 
 | | Structure of Helix Pomatia agglutinin with no ligands | | Descriptor: | HELIX POMATIA AGGLUTININ | | Authors: | Sanchez, J.-F, Lescar, J, Chazalet, V, Audfray, A, Gautier, C, Gagnon, J, Breton, C, Imberty, A, Mitchell, E.P. | | Deposit date: | 2006-02-03 | | Release date: | 2006-05-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Biochemical and Structural Analysis of Helix Pomatia Agglutinin. A Hexameric Lectin with a Novel Fold.
J.Biol.Chem., 281, 2006
|
|
8A5B
 
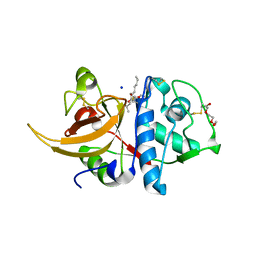 | | Crystal structure of human cathepsin L in complex with covalently bound MG-101 | | Descriptor: | Calpain Inhibitor I, Cathepsin L, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Falke, S, Lieske, J, Guenther, S, Reinke, P.Y.A, Ewert, W, Loboda, J, Karnicar, K, Usenik, A, Lindic, N, Sekirnik, A, Chapman, H.N, Hinrichs, W, Turk, D, Meents, A. | | Deposit date: | 2022-06-14 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Elucidation and Antiviral Activity of Covalent Cathepsin L Inhibitors.
J.Med.Chem., 67, 2024
|
|
4IXR
 
 | | RT fs X-ray diffraction of Photosystem II, first illuminated state | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Alonso-Mori, R, Tran, R, Hattne, J, Gildea, R.J, Echols, N, Gloeckner, C, Hellmich, J, Laksmono, H, Sierra, R.G, Lassalle-Kaiser, B, Koroidov, S, Lampe, A, Han, G, Gul, S, DiFiore, D, Milathianaki, D, Fry, A.R, Miahnahri, A, Schafer, D.W, Messerschmidt, M, Seibert, M.M, Koglin, J.E, Sokaras, D, Weng, T.-C, Sellberg, J, Latimer, M.J, Grosse-Kunstleve, R.W, Zwart, P.H, White, W.E, Glatzel, P, Adams, P.D, Bogan, M.J, Williams, G.J, Boutet, S, Messinger, J, Zouni, A, Sauter, N.K, Yachandra, V.K, Bergmann, U, Yano, J. | | Deposit date: | 2013-01-27 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (5.9 Å) | | Cite: | Simultaneous femtosecond X-ray spectroscopy and diffraction of photosystem II at room temperature.
Science, 340, 2013
|
|
7QK7
 
 | | Crystal structure of the APO form of ALDH1A3 | | Descriptor: | Aldehyde dehydrogenase family 1 member A3, DI(HYDROXYETHYL)ETHER, GLYCEROL | | Authors: | Castellvi, A, Farres, J. | | Deposit date: | 2021-12-17 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.289 Å) | | Cite: | Structural and biochemical evidence that ATP inhibits the cancer biomarker human aldehyde dehydrogenase 1A3.
Commun Biol, 5, 2022
|
|
7QK8
 
 | | Crystal structure of the ALDH1A3-NAD+ complex | | Descriptor: | Aldehyde dehydrogenase family 1 member A3, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Castellvi, A, Farres, J. | | Deposit date: | 2021-12-17 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.893 Å) | | Cite: | Structural and biochemical evidence that ATP inhibits the cancer biomarker human aldehyde dehydrogenase 1A3.
Commun Biol, 5, 2022
|
|
7P77
 
 | | SARS-CoV-2 spike protein in complex with sybody#15 and sybody#68 in a 3up conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(5-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Walter, J.D, Hutter, C.A.J, Garaeva, A.A, Scherer, M, Zimmermann, I, Wyss, M, Rheinberger, J, Ruedin, Y, Earp, J.C, Egloff, P, Sorgenfrei, M, Huerlimann, L.M, Gonda, I, Meier, G, Remm, S, Thavarasah, S, Zimmer, G, Slotboom, D.J, Paulino, C, Plattet, P, Seeger, M.A. | | Deposit date: | 2021-07-19 | | Release date: | 2021-08-04 | | Last modified: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Biparatopic sybodies neutralize SARS-CoV-2 variants of concern and mitigate drug resistance.
Embo Rep., 23, 2022
|
|
7QK9
 
 | | Crystal structure of the ALDH1A3-ATP complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Aldehyde dehydrogenase family 1 member A3, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Castellvi, A, Farres, J. | | Deposit date: | 2021-12-17 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.782 Å) | | Cite: | Structural and biochemical evidence that ATP inhibits the cancer biomarker human aldehyde dehydrogenase 1A3.
Commun Biol, 5, 2022
|
|
2C8Q
 
 | | insuline(1sec) and UV laser excited fluorescence | | Descriptor: | INSULIN A CHAIN, INSULIN B CHAIN | | Authors: | Vernede, X, Lavault, B, Ohana, J, Nurizzo, D, Joly, J, Jacquamet, L, Felisaz, F, Cipriani, F, Bourgeois, D. | | Deposit date: | 2005-12-06 | | Release date: | 2006-03-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Uv Laser-Excited Fluorescence as a Tool for the Visualization of Protein Crystals Mounted in Loops.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
7P7A
 
 | | SARS-CoV-2 spike protein in complex with sybody#68 in a 2up/1flexible conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(5-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Walter, J.D, Hutter, C.A.J, Garaeva, A.A, Scherer, M, Zimmermann, I, Wyss, M, Rheinberger, J, Ruedin, Y, Earp, J.C, Egloff, P, Sorgenfrei, M, Huerlimann, L.M, Gonda, I, Meier, G, Remm, S, Thavarasah, S, Zimmer, G, Slotboom, D.J, Paulino, C, Plattet, P, Seeger, M.A. | | Deposit date: | 2021-07-19 | | Release date: | 2021-08-04 | | Last modified: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (4.76 Å) | | Cite: | Biparatopic sybodies neutralize SARS-CoV-2 variants of concern and mitigate drug resistance.
Embo Rep., 23, 2022
|
|
7P78
 
 | | SARS-CoV-2 spike protein in complex with sybody#15 and sybody#68 in a 1up/1up-out/1down conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(5-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Walter, J.D, Hutter, C.A.J, Garaeva, A.A, Scherer, M, Zimmermann, I, Wyss, M, Rheinberger, J, Ruedin, Y, Earp, J.C, Egloff, P, Sorgenfrei, M, Huerlimann, L.M, Gonda, I, Meier, G, Remm, S, Thavarasah, S, Zimmer, G, Slotboom, D.J, Paulino, C, Plattet, P, Seeger, M.A. | | Deposit date: | 2021-07-19 | | Release date: | 2021-08-04 | | Last modified: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Biparatopic sybodies neutralize SARS-CoV-2 variants of concern and mitigate drug resistance.
Embo Rep., 23, 2022
|
|
6TQR
 
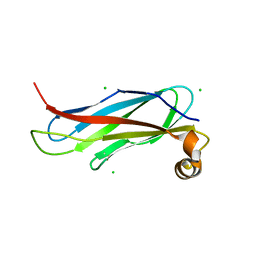 | | The crystal structure of the MSP domain of human VAP-A in complex with the Phospho-FFAT motif of STARD3. | | Descriptor: | CHLORIDE ION, StAR-related lipid transfer protein 3, Vesicle-associated membrane protein-associated protein A | | Authors: | McEwen, A.G, Poussin-Courmontagne, P, Di Mattia, T, Wendling, C, Cavarelli, J, Tomasetto, C, Alpy, F. | | Deposit date: | 2019-12-17 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | FFAT motif phosphorylation controls formation and lipid transfer function of inter-organelle contacts.
Embo J., 39, 2020
|
|
6PUH
 
 | | Structure of human MAIT A-F7 TCR in complex with human MR1-Ribityl-less | | Descriptor: | 6-methyl-5-[(1E)-3-oxobut-1-en-1-yl]pyrimidine-2,4(1H,3H)-dione, Beta-2-microglobulin, Human TCR alpha chain, ... | | Authors: | Awad, W, Keller, A.N, Rossjohn, J. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The molecular basis underpinning the potency and specificity of MAIT cell antigens.
Nat.Immunol., 21, 2020
|
|
7TPR
 
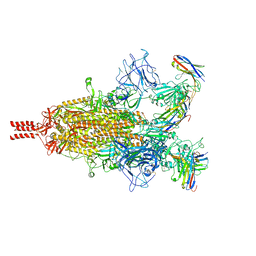 | | Camel nanobodies 7A3 and 8A2 broadly neutralize SARS-CoV-2 variants | | Descriptor: | Nanobody 7A3, Nanobody 8A2, Spike glycoprotein | | Authors: | Butay, K.J, Zhu, J, Dandey, V.P, Hong, J, Kwon, H.J, Chen, C.Z, Duan, Z, Li, D, Ren, H, Liang, T, Martin, N, Esposito, D, Ortega-Rodriguez, U, Xu, M, Xie, H, Ho, M, Cachau, R, Borgnia, M.J. | | Deposit date: | 2022-01-25 | | Release date: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (2.39 Å) | | Cite: | Camel nanobodies broadly neutralize SARS-CoV-2 variants
bioRxiv, 2021
|
|
2CLX
 
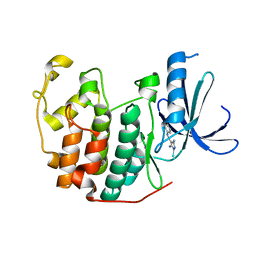 | | 4-Arylazo-3,5-diamino-1H-pyrazole CDK Inhibitors: SAR Study, Crystal Structure in Complex with CDK2, Selectivity, and Cellular Effects | | Descriptor: | 4-[(E)-(3,5-DIAMINO-1H-PYRAZOL-4-YL)DIAZENYL]PHENOL, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Krystof, V, Cankar, P, Frysova, I, Slouka, J, Kontopidis, G, Dzubak, P, Hajduch, M, Deazevedo, W.F, Paprskarova, M, Orsag, M, Rolcik, J, Latr, A, Fischer, P.M, Strnad, M. | | Deposit date: | 2006-05-02 | | Release date: | 2006-11-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 4-Arylazo-3,5-Diamino-1H-Pyrazole Cdk Inhibitors: Sar Study, Crystal Structure in Complex with Cdk2, Selectivity, and Cellular Effects
J.Med.Chem., 49, 2006
|
|
7P79
 
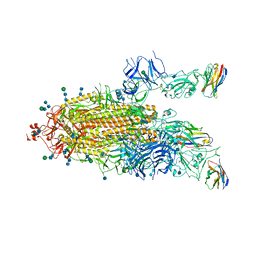 | | SARS-CoV-2 spike protein in complex with sybodyb#15 in a 1up/1up-out/1down conformation. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(5-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Walter, J.D, Hutter, C.A.J, Garaeva, A.A, Scherer, M, Zimmermann, I, Wyss, M, Rheinberger, J, Ruedin, Y, Earp, J.C, Egloff, P, Sorgenfrei, M, Huerlimann, L.M, Gonda, I, Meier, G, Remm, S, Thavarasah, S, Zimmer, G, Slotboom, D.J, Paulino, C, Plattet, P, Seeger, M.A. | | Deposit date: | 2021-07-19 | | Release date: | 2021-08-04 | | Last modified: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Biparatopic sybodies neutralize SARS-CoV-2 variants of concern and mitigate drug resistance.
Embo Rep., 23, 2022
|
|
8JB0
 
 | |
8A1U
 
 | | Sodium pumping NADH-quinone oxidoreductase with substrates NADH and Q2 | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Hau, J.-L, Kaltwasser, S, Vonck, J, Fritz, G, Steuber, J. | | Deposit date: | 2022-06-02 | | Release date: | 2023-09-20 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Conformational coupling of redox-driven Na + -translocation in Vibrio cholerae NADH:quinone oxidoreductase.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7P7B
 
 | | SARS-CoV-2 spike protein in complex with sybody no68 in a 1up/2down conformation | | Descriptor: | Spike glycoprotein | | Authors: | Walter, J.D, Hutter, C.A.J, Garaeva, A.A, Scherer, M, Zimmermann, I, Wyss, M, Rheinberger, J, Ruedin, Y, Earp, J.C, Egloff, P, Sorgenfrei, M, Huerlimann, L.M, Gonda, I, Meier, G, Remm, S, Thavarasah, S, Zimmer, G, Slotboom, D.J, Paulino, C, Plattet, P, Seeger, M.A. | | Deposit date: | 2021-07-19 | | Release date: | 2021-08-04 | | Last modified: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Biparatopic sybodies neutralize SARS-CoV-2 variants of concern and mitigate drug resistance.
Embo Rep., 23, 2022
|
|
7OZV
 
 | | SARS-CoV-2 RdRp with Molnupiravir/ NHC in the template strand base-paired with G | | Descriptor: | Non-structural protein 7, Non-structural protein 8, Product RNA, ... | | Authors: | Kabinger, F, Stiller, C, Schmitzova, J, Dienemann, C, Kokic, G, Hillen, H.S, Hoebartner, C, Cramer, P. | | Deposit date: | 2021-06-28 | | Release date: | 2021-08-18 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of molnupiravir-induced SARS-CoV-2 mutagenesis.
Nat.Struct.Mol.Biol., 28, 2021
|
|
