6NNR
 
 | |
1GI6
 
 | | A NOVEL SERINE PROTEASE INHIBITION MOTIF INVOLVING A MULTI-CENTERED SHORT HYDROGEN BONDING NETWORK AT THE ACTIVE SITE | | 分子名称: | 2-(2-HYDROXY-PHENYL)-1H-INDOLE-5-CARBOXAMIDINE, BETA-TRYPSIN, CALCIUM ION, ... | | 著者 | Katz, B.A, Elrod, K, Luong, C, Rice, M, Mackman, R.L, Sprengeler, P.A, Spencer, J, Hatayte, J, Janc, J, Link, J, Litvak, J, Rai, R, Rice, K, Sideris, S, Verner, E, Young, W. | | 登録日 | 2001-01-22 | | 公開日 | 2002-01-22 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | A novel serine protease inhibition motif involving a multi-centered short hydrogen bonding network at the active site.
J.Mol.Biol., 307, 2001
|
|
7QUU
 
 | | Red1-Iss10 complex | | 分子名称: | NURS complex subunit red1, Uncharacterized protein C7D4.14c | | 著者 | Mackereth, C.D, Kadlec, J, Laroussi, H. | | 登録日 | 2022-01-18 | | 公開日 | 2022-09-07 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural analysis of Red1 as a conserved scaffold of the RNA-targeting MTREC/PAXT complex.
Nat Commun, 13, 2022
|
|
6BXH
 
 | | Menin in complex with MI-853 | | 分子名称: | 1-{2-[4-(fluoroacetyl)piperazin-1-yl]ethyl}-4-methyl-5-[(4-{[6-(2,2,2-trifluoroethyl)thieno[2,3-d]pyrimidin-4-yl]amino}piperidin-1-yl)methyl]-1H-indole-2-carbonitrile, DIMETHYL SULFOXIDE, Menin, ... | | 著者 | Borkin, D, Klossowski, S, Pollock, J, Linhares, B, Cierpicki, T, Grembecka, J. | | 登録日 | 2017-12-18 | | 公開日 | 2018-11-28 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.445 Å) | | 主引用文献 | Menin in complex with MI-853
To Be Published
|
|
6HGT
 
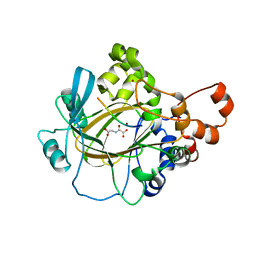 | |
2VML
 
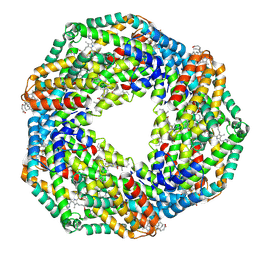 | |
2UUO
 
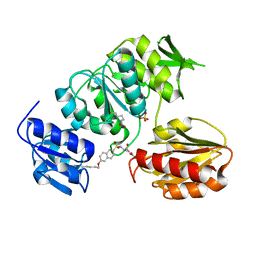 | | Crystal structure of MurD ligase in complex with D-Glu containing sulfonamide inhibitor | | 分子名称: | N-{[6-(PENTYLOXY)NAPHTHALEN-2-YL]SULFONYL}-D-GLUTAMIC ACID, SULFATE ION, UDP-N-ACETYLMURAMOYLALANINE--D-GLUTAMATE LIGASE | | 著者 | Humljan, J, Kotnik, M, Contreras-Martel, C, Blanot, D, Urleb, U, Dessen, A, Solmajer, T, Gobec, S. | | 登録日 | 2007-03-06 | | 公開日 | 2008-03-25 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Novel naphthalene-N-sulfonyl-D-glutamic acid derivatives as inhibitors of MurD, a key peptidoglycan biosynthesis enzyme.
J. Med. Chem., 51, 2008
|
|
7QV7
 
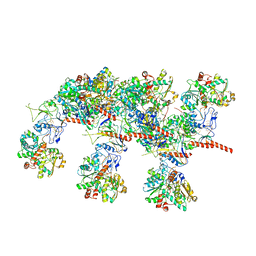 | | Cryo-EM structure of Hydrogen-dependent CO2 reductase. | | 分子名称: | Hydrogen dependent carbon dioxide reductase subunit FdhF, Hydrogen dependent carbon dioxide reductase subunit HycB3, Hydrogen dependent carbon dioxide reductase subunit HycB4, ... | | 著者 | Dietrich, H.M, Righetto, R.D, Kumar, A, Wietrzynski, W, Schuller, S.K, Trischler, R, Wagner, J, Schwarz, F.M, Engel, B.D, Mueller, V, Schuller, J.M. | | 登録日 | 2022-01-19 | | 公開日 | 2022-07-06 | | 最終更新日 | 2022-08-10 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Membrane-anchored HDCR nanowires drive hydrogen-powered CO 2 fixation.
Nature, 607, 2022
|
|
7QDQ
 
 | | Crystal Structure of HDM2 in complex with Caylin-1 | | 分子名称: | CHLORIDE ION, Caylin-1, DIMETHYL SULFOXIDE, ... | | 著者 | Finke, A.D, Walti, M.A, Marsh, M.E, Orts, J. | | 登録日 | 2021-11-29 | | 公開日 | 2022-10-05 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.26 Å) | | 主引用文献 | Elucidation of a nutlin-derivative-HDM2 complex structure at the interaction site by NMR molecular replacement: A straightforward derivation
J Magn Reson Open, 10-11, 2022
|
|
2V1C
 
 | |
7QAA
 
 | | Crystal structure of RARalpha/RXRalpha ligand binding domain heterodimer in complex with BMS614 and oleic acid | | 分子名称: | 4-[(4,4-DIMETHYL-1,2,3,4-TETRAHYDRO-[1,2']BINAPTHALENYL-7-CARBONYL)-AMINO]-BENZOIC ACID, Isoform Alpha-1-deltaBC of Retinoic acid receptor alpha, OLEIC ACID, ... | | 著者 | le Maire, A, Vivat, V, Guee, L, Blanc, P, Malosse, C, Chamot-Rooke, J, Germain, P, Bourguet, w. | | 登録日 | 2021-11-16 | | 公開日 | 2022-10-05 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.76 Å) | | 主引用文献 | Design and in vitro characterization of RXR variants as tools to investigate the biological role of endogenous rexinoids.
J.Mol.Endocrinol., 69, 2022
|
|
4PJ9
 
 | | Structure of human MR1-5-OP-RU in complex with human MAIT TRAJ20 TCR | | 分子名称: | 1-deoxy-1-({2,6-dioxo-5-[(E)-propylideneamino]-1,2,3,6-tetrahydropyrimidin-4-yl}amino)-D-ribitol, Beta-2-microglobulin, GLYCEROL, ... | | 著者 | Birkinshaw, R.W, Rossjohn, J. | | 登録日 | 2014-05-12 | | 公開日 | 2014-07-02 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A molecular basis underpinning the T cell receptor heterogeneity of mucosal-associated invariant T cells.
J.Exp.Med., 211, 2014
|
|
6TET
 
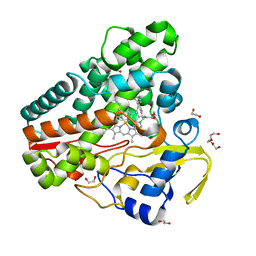 | | The structure of CYP121 in complex with inhibitor L21 | | 分子名称: | 1,2-ETHANEDIOL, 1-[(~{E})-3-[4-(4-fluorophenyl)phenyl]prop-2-enyl]imidazole, Mycocyclosin synthase, ... | | 著者 | Adam, S, Koehnke, J. | | 登録日 | 2019-11-12 | | 公開日 | 2021-05-26 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.49986887 Å) | | 主引用文献 | Structure-Activity Relationship and Mode-Of-Action Studies Highlight 1-(4-Biphenylylmethyl)-1H-imidazole-Derived Small Molecules as Potent CYP121 Inhibitors.
Chemmedchem, 16, 2021
|
|
4PJG
 
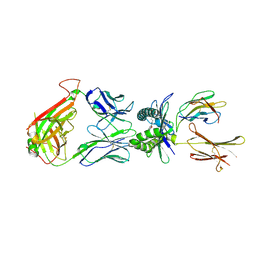 | |
7QY6
 
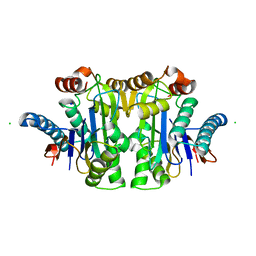 | | Structure of E.coli Class 2 L-asparaginase EcAIII, wild type (WT EcAIII) | | 分子名称: | Beta-aspartyl-peptidase, CHLORIDE ION, Isoaspartyl peptidase, ... | | 著者 | Loch, J.I, Klonecka, A, Kadziolka, K, Bonarek, P, Barciszewski, J, Imiolczyk, B, Brzezinski, K, Jaskolski, M. | | 登録日 | 2022-01-27 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structural and biophysical studies of new L-asparaginase variants: lessons from random mutagenesis of the prototypic Escherichia coli Ntn-amidohydrolase.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
6TEV
 
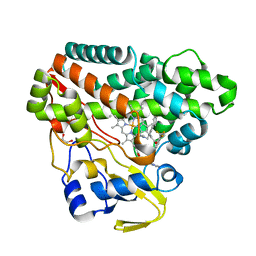 | | The structure of CYP121 in complex with inhibitor L44 | | 分子名称: | 1,2-ETHANEDIOL, 1-[[4-[4-(trifluoromethyl)phenyl]phenyl]methyl]imidazole, Mycocyclosin synthase, ... | | 著者 | Adam, S, Koehnke, J. | | 登録日 | 2019-11-12 | | 公開日 | 2021-05-26 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.70001268 Å) | | 主引用文献 | Structure-Activity Relationship and Mode-Of-Action Studies Highlight 1-(4-Biphenylylmethyl)-1H-imidazole-Derived Small Molecules as Potent CYP121 Inhibitors.
Chemmedchem, 16, 2021
|
|
4PJX
 
 | | Structure of human MR1-Ac-6-FP in complex with human MAIT C-A11 TCR | | 分子名称: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-2-microglobulin, CHLORIDE ION, ... | | 著者 | Birkinshaw, R.W, Rossjohn, J. | | 登録日 | 2014-05-12 | | 公開日 | 2014-07-02 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | A molecular basis underpinning the T cell receptor heterogeneity of mucosal-associated invariant T cells.
J.Exp.Med., 211, 2014
|
|
6C96
 
 | | Cryo-EM structure of mouse TPC1 channel in the apo state | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SODIUM ION, ... | | 著者 | She, J, Guo, J, Chen, Q, Bai, X, Jiang, Y. | | 登録日 | 2018-01-25 | | 公開日 | 2018-04-04 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural insights into the voltage and phospholipid activation of the mammalian TPC1 channel.
Nature, 556, 2018
|
|
6TE7
 
 | | The structure of CYP121 in complex with inhibitor S2 | | 分子名称: | 1,2-ETHANEDIOL, 2-chloranyl-4-[4-[(1~{R})-1-imidazol-1-ylprop-2-enyl]phenyl]phenol, Mycocyclosin synthase, ... | | 著者 | Adam, S, Koehnke, J. | | 登録日 | 2019-11-11 | | 公開日 | 2021-05-26 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.50001824 Å) | | 主引用文献 | Structure-Activity Relationship and Mode-Of-Action Studies Highlight 1-(4-Biphenylylmethyl)-1H-imidazole-Derived Small Molecules as Potent CYP121 Inhibitors.
Chemmedchem, 16, 2021
|
|
4PLB
 
 | | Crystal Structure of S.A. gyrase-AM8191 complex | | 分子名称: | 6-[({(1r,4S)-1-[(1S)-2-(3-fluoro-6-methoxy-1,5-naphthyridin-4-yl)-1-hydroxyethyl]-2-oxabicyclo[2.2.2]oct-4-yl}amino)methyl]-2H-pyrido[3,2-b][1,4]oxazin-3(4H)-one, Chimera protein of DNA gyrase subunits B and A, DNA (5'-D(P*AP*GP*CP*CP*GP*TP*AP*GP*GP*GP*CP*CP*CP*TP*AP*CP*GP*GP*CP*T)-3'), ... | | 著者 | Lu, J, Patel, S, Soisson, S. | | 登録日 | 2014-05-16 | | 公開日 | 2014-06-18 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.69 Å) | | 主引用文献 | Oxabicyclooctane-linked novel bacterial topoisomerase inhibitors as broad spectrum antibacterial agents.
Acs Med.Chem.Lett., 5, 2014
|
|
5N8W
 
 | | CRYSTAL STRUCTURE OF STREPTAVIDIN with D-amino acid containing peptide GGwhdeatwkpG | | 分子名称: | GLY-GLY-DTR-DHI-DAS-DGL-DAL-DTH-DTR-DLY, Streptavidin | | 著者 | Lyamichev, V, Goodrich, L, Sullivan, E, Bannen, R, Benz, J, Albert, T, Patel, J. | | 登録日 | 2017-02-24 | | 公開日 | 2017-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Stepwise Evolution Improves Identification of Diverse Peptides Binding to a Protein Target.
Sci Rep, 7, 2017
|
|
6HLF
 
 | | X-ray structure of Lactobacillus brevis alcohol dehydrogenase mutant - K32A | | 分子名称: | MAGNESIUM ION, MANGANESE (II) ION, R-specific alcohol dehydrogenase | | 著者 | Hermann, J, Nowotny, P, Schneider, S, Hekmat, D, Weuster-Botz, D. | | 登録日 | 2018-09-11 | | 公開日 | 2018-12-12 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Rational Crystal Contact Engineering of Lactobacillus brevis Alcohol Dehydrogenase To Promote Technical Protein Crystallization
Cryst.Growth Des., 2019
|
|
2OC1
 
 | | Structure of the HCV NS3/4A Protease Inhibitor CVS4819 | | 分子名称: | (2S)-({N-[(3S)-3-({N-[(2S,4E)-2-ISOPROPYL-7-METHYLOCT-4-ENOYL]-L-LEUCYL}AMINO)-2-OXOHEXANOYL]GLYCYL}AMINO)(PHENYL)ACETI C ACID, Hepatitis C virus, ZINC ION | | 著者 | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | 登録日 | 2006-12-20 | | 公開日 | 2007-07-31 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
6TI5
 
 | | A New Structural Model of Abeta(1-40) Fibrils | | 分子名称: | Amyloid-beta precursor protein | | 著者 | Bertini, I, Gonnelli, L, Luchinat, C, Mao, J, Nesi, A. | | 登録日 | 2019-11-21 | | 公開日 | 2020-07-22 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLID-STATE NMR | | 主引用文献 | Mixing A beta (1-40) and A beta (1-42) peptides generates unique amyloid fibrils.
Chem.Commun.(Camb.), 56, 2020
|
|
7QYM
 
 | | Structure of E.coli Class 2 L-asparaginase EcAIII, mutant RDM1-18 (R207V, D210P, S211W) | | 分子名称: | Beta-aspartyl-peptidase, CHLORIDE ION, Isoaspartyl peptidase, ... | | 著者 | Loch, J.I, Klonecka, A, Kadziolka, K, Bonarek, P, Barciszewski, J, Imiolczyk, B, Brzezinski, K, Jaskolski, M. | | 登録日 | 2022-01-28 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Structural and biophysical studies of new L-asparaginase variants: lessons from random mutagenesis of the prototypic Escherichia coli Ntn-amidohydrolase.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
