5TKN
 
 | |
4NM6
 
 | | Crystal structure of TET2-DNA complex | | Descriptor: | 5'-D(*AP*CP*CP*AP*CP*(5CM)P*GP*GP*TP*GP*GP*T)-3', FE (II) ION, Methylcytosine dioxygenase TET2, ... | | Authors: | Hu, L, Li, Z, Cheng, J, Rao, Q, Gong, W, Liu, M, Wang, P, Xu, Y. | | Deposit date: | 2013-11-14 | | Release date: | 2013-12-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.026 Å) | | Cite: | Crystal Structure of TET2-DNA Complex: Insight into TET-Mediated 5mC Oxidation.
Cell(Cambridge,Mass.), 155, 2013
|
|
1FAP
 
 | | THE STRUCTURE OF THE IMMUNOPHILIN-IMMUNOSUPPRESSANT FKBP12-RAPAMYCIN COMPLEX INTERACTING WITH HUMAN FRAP | | Descriptor: | FK506-BINDING PROTEIN, FRAP, RAPAMYCIN IMMUNOSUPPRESSANT DRUG | | Authors: | Choi, J, Chen, J, Schreiber, S.L, Clardy, J. | | Deposit date: | 1996-03-15 | | Release date: | 1997-07-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the FKBP12-rapamycin complex interacting with the binding domain of human FRAP.
Science, 273, 1996
|
|
8BOC
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 19 | | Descriptor: | Ephrin type-A receptor 2, ~{N}-[3,5-bis(trifluoromethyl)phenyl]-4-methyl-3-[(1-methyl-6-pyridin-3-yl-pyrazolo[3,4-d]pyrimidin-4-yl)amino]benzamide | | Authors: | Linhard, V, Witt, K, Gande, S, Wollenhaupt, J, Lennartz, F, Weiss, M.S, Schwalbe, H. | | Deposit date: | 2022-11-15 | | Release date: | 2023-11-29 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Optimization of the Lead Compound NVP-BHG712 as a Colorectal Cancer Inhibitor.
Chemistry, 29, 2023
|
|
6O7K
 
 | | 30S initiation complex | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Frank, J, Gonzalez Jr, R.L, kaledhonkar, S, Fu, Z, Caban, K, Li, W, Chen, B, Sun, M. | | Deposit date: | 2019-03-08 | | Release date: | 2019-05-29 | | Last modified: | 2020-01-29 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Late steps in bacterial translation initiation visualized using time-resolved cryo-EM.
Nature, 570, 2019
|
|
4IX1
 
 | | Crystal structure of hypothetical protein OPAG_01669 from Rhodococcus Opacus PD630, Target 016205 | | Descriptor: | PHOSPHATE ION, hypothetical protein | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Zenchek, W, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-01-24 | | Release date: | 2013-02-06 | | Last modified: | 2013-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of hypothetical protein OPAG_01669 from Rhodococcus Opacus PD630, Target 016205
To be Published
|
|
2XEM
 
 | | Induced-fit and allosteric effects upon polyene binding revealed by crystal structures of the Dynemicin thioesterase | | Descriptor: | (3E,5E,7E,9E,11E,13E)-pentadeca-3,5,7,9,11,13-hexaen-2-one, DYNE7 | | Authors: | Liew, C.W, Sharff, A, Kotaka, M, Kong, R, Bricogne, G, Liang, Z.X, Lescar, J. | | Deposit date: | 2010-05-17 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Induced-Fit Upon Ligand Binding Revealed by Crystal Structures of the Hot-Dog Fold Thioesterase in Dynemicin Biosynthesis.
J.Mol.Biol., 404, 2010
|
|
5YLE
 
 | | MCR-1 complex with ethanolamine (ETA) | | Descriptor: | ETHANOLAMINE, Probable phosphatidylethanolamine transferase Mcr-1, ZINC ION | | Authors: | Wei, P.C, Song, G.J, Shi, M.Y, Zhou, Y.F, Liu, Y, Lei, J, Chen, P, Yin, L. | | Deposit date: | 2017-10-17 | | Release date: | 2017-11-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substrate analog interaction with MCR-1 offers insight into the rising threat of the plasmid-mediated transferable colistin resistance.
FASEB J., 32, 2018
|
|
2C8D
 
 | | Structure of the ARTT motif Q212A mutant C3bot1 Exoenzyme (Free state, crystal form I) | | Descriptor: | MONO-ADP-RIBOSYLTRANSFERASE C3, SULFATE ION | | Authors: | Stura, E.A, Menetrey, J, Flatau, G, Boquet, P, Menez, A. | | Deposit date: | 2005-12-03 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for the Nad-Hydrolysis Mechanism and the Artt-Loop Plasticity of C3 Exoenzymes.
Protein Sci., 17, 2008
|
|
2OYR
 
 | | Crystal Structure of UPF0341 Protein (yhiQ) from Shigella flexneri in complex with S-Adenosyl Homocysteine, Northeast Structural Genomics Target SfR275 | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, UPF0341 protein yhiQ | | Authors: | Forouhar, F, Su, M, Seetharaman, J, Janjua, H, Fang, Y, Xiao, R, Baran, M.C, Liu, J, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-02-22 | | Release date: | 2007-03-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of UPF0341 Protein (yhiQ) from Shigella flexneri in complex with S-Adenosyl Homocysteine, Northeast Structural Genomics Target SfR275
To be Published
|
|
2IAX
 
 | | Crystal structure of squid ganglion DFPase D232S mutant | | Descriptor: | CALCIUM ION, Diisopropylfluorophosphatase | | Authors: | Katsemi, V, Luecke, C, Koepke, J, Loehr, F, Maurer, S, Fritzsch, G, Rueterjans, H. | | Deposit date: | 2006-09-08 | | Release date: | 2006-09-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Mutational and structural studies of the diisopropylfluorophosphatase from Loligo vulgaris shed new light on the catalytic mechanism of the enzyme
Biochemistry, 44, 2005
|
|
7JW2
 
 | | Crystal structure of Aedes aegypti Nibbler EXO domain | | Descriptor: | Exonuclease mut-7 homolog | | Authors: | Xie, W, Sowemimo, I, Hayashi, R, Wang, J, Brennecke, J, Ameres, S.L, Patel, D.J. | | Deposit date: | 2020-08-24 | | Release date: | 2021-01-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-function analysis of microRNA 3'-end trimming by Nibbler.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5TBD
 
 | | Crystal Structure of anti-MSP2 Fv fragment (mAb4D11) in complex with 3D7-MSP2 215-222 | | Descriptor: | Fv fragment (mAb4D1) heavy chain, Merozoite surface protein 2 | | Authors: | Seow, J, Morales, R.A.V, MacRaild, C.A, Bankala, K, Drinkwater, N, Dingjan, T, Jaipuria, G, Wilde, K, Anders, R.F, Atreya, H.S, Christ, D, McGowan, S, Norton, R.S. | | Deposit date: | 2016-09-12 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Characterisation of a Key Epitope in the Conserved C-Terminal Domain of the Malaria Vaccine Candidate MSP2.
J. Mol. Biol., 429, 2017
|
|
5TBS
 
 | | Crystal Structure of Isoform 2 of Purine Nucleoside Phosphorylase complexed with adenine | | Descriptor: | ADENINE, DIMETHYL SULFOXIDE, Purine nucleoside phosphorylase | | Authors: | Faheem, M, Torini, J.R, Romanello, L, Brandao-Neto, J, Pereira, H.M. | | Deposit date: | 2016-09-13 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The molecular structure of Schistosoma mansoni PNP isoform 2 provides insights into the nucleoside selectivity of PNPs.
PLoS ONE, 13, 2018
|
|
5TBT
 
 | | Crystal Structure of Isoform 2 of Purine Nucleoside Phosphorylase complexed with Cytidine | | Descriptor: | 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, Purine nucleoside phosphorylase, SULFATE ION | | Authors: | Faheem, M, Torini, J.R, Romanello, L, Brandao-Neto, J, Pereira, H.M. | | Deposit date: | 2016-09-13 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | The molecular structure of Schistosoma mansoni PNP isoform 2 provides insights into the nucleoside selectivity of PNPs.
PLoS ONE, 13, 2018
|
|
2C8E
 
 | | Structure of the ARTT motif E214N mutant C3bot1 Exoenzyme (Free state, crystal form III) | | Descriptor: | MONO-ADP-RIBOSYLTRANSFERASE C3, SULFATE ION | | Authors: | Stura, E.A, Menetrey, J, Flatau, G, Boquet, P, Menez, A. | | Deposit date: | 2005-12-03 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis for the Nad-Hydrolysis Mechanism and the Artt-Loop Plasticity of C3 Exoenzymes.
Protein Sci., 17, 2008
|
|
2P9M
 
 | | Crystal structure of conserved hypothetical protein MJ0922 from Methanocaldococcus jannaschii DSM 2661 | | Descriptor: | Hypothetical protein MJ0922 | | Authors: | Zhao, M, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, Zhu, J, Swindell II, J.T, Chen, L, Fu, Z.-Q, Charz, J, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-26 | | Release date: | 2007-07-03 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure of conserved hypothetical protein MJ0922 from Methanocaldococcus jannaschii DSM 2661
To be Published
|
|
2IAR
 
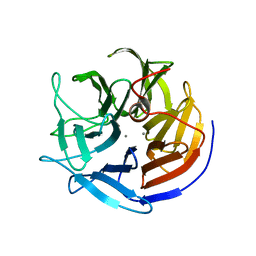 | | Crystal structure of squid ganglion DFPase W244H mutant | | Descriptor: | CALCIUM ION, Diisopropylfluorophosphatase | | Authors: | Scharff, E.I, Koepke, J, Fritzsch, G, Luecke, C, Rueterjans, H. | | Deposit date: | 2006-09-08 | | Release date: | 2006-09-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of diisopropylfluorophosphatase from Loligo vulgaris
Structure, 9, 2001
|
|
2C8B
 
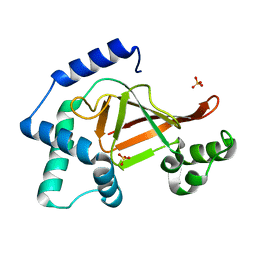 | | Structure of the ARTT motif Q212A mutant C3bot1 Exoenzyme (Free state, crystal form II) | | Descriptor: | MONO-ADP-RIBOSYLTRANSFERASE C3, SULFATE ION | | Authors: | Stura, E.A, Menetrey, J, Flatau, G, Boquet, P, Menez, A. | | Deposit date: | 2005-12-03 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for the Nad-Hydrolysis Mechanism and the Artt-Loop Plasticity of C3 Exoenzymes.
Protein Sci., 17, 2008
|
|
8BRF
 
 | |
7ULY
 
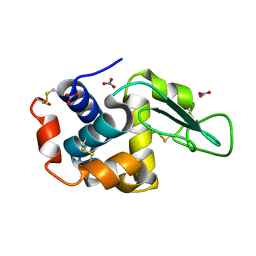 | | MicroED structure of triclinic lysozyme | | Descriptor: | Lysozyme C, NITRATE ION | | Authors: | Clabbers, M.T.B, Martynowycz, M.W, Hattne, J, Gonen, T. | | Deposit date: | 2022-04-05 | | Release date: | 2023-03-15 | | Last modified: | 2024-10-09 | | Method: | ELECTRON CRYSTALLOGRAPHY (0.87 Å) | | Cite: | Hydrogens and hydrogen-bond networks in macromolecular MicroED data.
J Struct Biol X, 6, 2022
|
|
4GF0
 
 | | Crystal structure of glutahtione transferase homolog from sulfitobacter, TARGET EFI-501084, with bound glutathione | | Descriptor: | CHLORIDE ION, GLUTATHIONE, Glutathione S-transferase | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-08-02 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of glutahtione transferase homolog from sulfitobacter, TARGET EFI-501084, with bound glutathione
To be Published
|
|
4A2J
 
 | | PR X-Ray structures in agonist conformations reveal two different mechanisms for partial agonism in 11beta-substituted steroids | | Descriptor: | 4-[(11BETA,17BETA)-17-METHOXY-17-(METHOXYMETHYL)-3-OXOESTRA-4,9-DIEN-11-YL]BENZALDEHYDE OXIME, PROGESTERONE RECEPTOR, SULFATE ION | | Authors: | Lusher, S.J, Raaijmakers, H.C.A, Bosch, R, Vu-Pham, D, McGuire, R, Oubrie, A, de Vlieg, J. | | Deposit date: | 2011-09-27 | | Release date: | 2012-04-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structures of progesterone receptor ligand binding domain in its agonist state reveal differing mechanisms for mixed profiles of 11 beta-substituted steroids.
J. Biol. Chem., 287, 2012
|
|
8BVA
 
 | | Crystal Structure of Mus musculus Protein Arginine Methyltransferase 2 in complex with RSF1_114-126 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[(~{E})-prop-1-enyl]oxolane-3,4-diol, L(+)-TARTARIC ACID, POTASSIUM ION, ... | | Authors: | Cura, V, Troffer-Charlier, N, Marechal, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2022-12-02 | | Release date: | 2023-12-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal Structure of Mus musculus Protein Arginine Methyltransferase 2 in complex with RSF1_114-126
To Be Published
|
|
6PFT
 
 | |
