9OB4
 
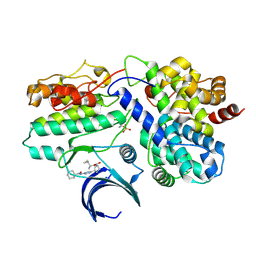 | |
4URV
 
 | | The crystal structure of H-Ras and SOS in complex with ligands | | Descriptor: | 4-(4-BROMOPHENYL)PIPERIDIN-4-OL, FORMIC ACID, GTPASE HRAS, ... | | Authors: | Winter, J.J.G, Anderson, M, Blades, K, Brassington, C, Breeze, A.L, Chresta, C, Embrey, K, Fairley, G, Faulder, P, Finlay, M.R.V, Kettle, J.G, Nowak, T, Overman, R, Patel, S.J, Perkins, P, Spadola, L, Tart, J, Tucker, J, Wrigley, G. | | Deposit date: | 2014-07-02 | | Release date: | 2015-03-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Small Molecule Binding Sites on the Ras:SOS Complex Can be Exploited for Inhibition of Ras Activation.
J.Med.Chem., 58, 2015
|
|
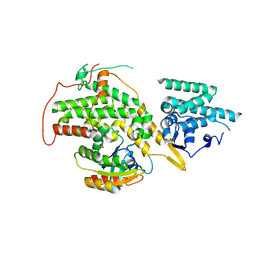
4UQK
 
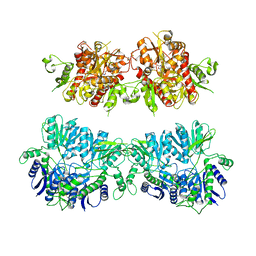 | | Electron density map of GluA2em in complex with quisqualate and LY451646 | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, GLUTAMATE RECEPTOR 2 | | Authors: | Meyerson, J.R, Kumar, J, Chittori, S, Rao, P, Pierson, J, Bartesaghi, A, Mayer, M.L, Subramaniam, S. | | Deposit date: | 2014-06-24 | | Release date: | 2014-08-13 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (16.4 Å) | | Cite: | Structural Mechanism of Glutamate Receptor Activation and Desensitization
Nature, 514, 2014
|
|
4V25
 
 | | VER-246608, a novel pan-isoform ATP competitive inhibitor of pyruvate dehydrogenase kinase, disrupts Warburg metabolism and induces context- dependent cytostasis in cancer cells | | Descriptor: | MAGNESIUM ION, N-(2-AMINOETHYL)-2-{3-CHLORO-4-[(4-ISOPROPYLBENZYL)OXY]PHENYL} ACETAMIDE, N-[4-(2-chloro-5-methylpyrimidin-4-yl)phenyl]-N-(4-{[(difluoroacetyl)amino]methyl}benzyl)-2,4-dihydroxybenzamide, ... | | Authors: | Moore, J.D, Staniszewska, A, Shaw, T, D'Alessandro, J, Davis, B, Surgenor, A, Baker, L, Matassova, N, Murray, J, Macias, A, Brough, P, Wood, M, Mahon, P.C. | | Deposit date: | 2014-10-06 | | Release date: | 2014-12-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | VER-246608, a novel pan-isoform ATP competitive inhibitor of pyruvate dehydrogenase kinase, disrupts Warburg metabolism and induces context-dependent cytostasis in cancer cells.
Oncotarget, 5, 2014
|
|
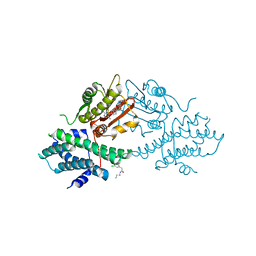
4UY1
 
 | | Novel pyrazole series of group X Secretory Phospholipase A2 (sPLA2-X) inhibitors | | Descriptor: | 5-(2,5-DIMETHYL-3-THIENYL)-1H-PYRAZOLE-3-CARBOXAMIDE, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sandmark, J, Oster, L, Hallberg, K, Bodin, C, Chen, H. | | Deposit date: | 2014-08-28 | | Release date: | 2014-10-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of a Novel Pyrazole Series of Group X Secreted Phospholipase A2 Inhibitor (Spla2X) Via Fragment Based Virtual Screening
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4UUU
 
 | | 1.7 A resolution structure of human cystathionine beta-synthase regulatory domain (del 516-525) in complex with SAM | | Descriptor: | 1,2-ETHANEDIOL, CYSTATHIONINE BETA-SYNTHASE, S-ADENOSYLMETHIONINE | | Authors: | Kopec, J, McCorvie, T.J, Fitzpatrick, F, Strain-Damerell, C, Froese, D.S, Tallant, C, Burgess-Brown, N, Arrowsmith, C, Edwards, A, Bountra, C, Yue, W.W. | | Deposit date: | 2014-07-31 | | Release date: | 2014-08-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Inter-Domain Communication of Human Cystathionine Beta Synthase: Structural Basis of S-Adenosyl-L-Methionine Activation.
J.Biol.Chem., 289, 2014
|
|
4W6E
 
 | | Human Tankyrase 1 with small molecule inhibitor | | Descriptor: | 2-(4-{6-[(3S)-3,4-dimethylpiperazin-1-yl]-4-methylpyridin-3-yl}phenyl)-8-(hydroxymethyl)quinazolin-4(3H)-one, Tankyrase-1, ZINC ION | | Authors: | Kazmirski, S.L, Johannes, J, Boriack-Sjodin, P.A, Howard, T. | | Deposit date: | 2014-08-20 | | Release date: | 2015-05-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Pyrimidinone nicotinamide mimetics as selective tankyrase and wnt pathway inhibitors suitable for in vivo pharmacology.
Acs Med.Chem.Lett., 6, 2015
|
|
4UR9
 
 | | Structure of ligand bound glycosylhydrolase | | Descriptor: | 4-ethoxyquinazoline, CALCIUM ION, O-(2-ACETAMIDO-2-DEOXY D-GLUCOPYRANOSYLIDENE) AMINO-N-PHENYLCARBAMATE, ... | | Authors: | Darby, J.F, Landstroem, J, Roth, C, He, Y, Schultz, M, Davies, G.J, Hubbard, R.E. | | Deposit date: | 2014-06-27 | | Release date: | 2015-02-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of Selective Small-Molecule Activators of a Bacterial Glycoside Hydrolase.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4UWF
 
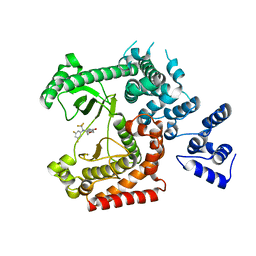 | | Discovery of (2S)-8-((3R)-3-Methylmorpholin-4-yl)-1-(3-methyl-2-oxo- butyl)-2-(trifluoromethyl)-3,4-dihydro-2H-pyrimido(1,2-a)pyrimidin-6- one: a Novel Potent and Selective Inhibitor of Vps34 for the Treatment of Solid Tumors | | Descriptor: | (8S)-9-[3,5-bis(fluoranyl)phenyl]-2-morpholin-4-yl-8-(trifluoromethyl)-7,8-dihydro-6H-pyrimido[1,2-a]pyrimidin-4-one, PHOSPHATIDYLINOSITOL 3-KINASE CATALYTIC SUBUNIT TYPE 3 | | Authors: | Pasquier, B, El-Ahmad, Y, Filoche-Romme, B, Dureuil, C, Fassy, F, Abecassis, P.Y, Mathieu, M, Bertrand, T, Benard, T, Barriere, C, ElBatti, S, Letallec, J.P, Sonnefraud, V, Brollo, M, Delbarre, L, Loyau, V, Pilorge, F, Bertin, L, Richepin, P, Arigon, J, Labrosse, J.R, Clement, J, Durand, F, Combet, R, Perraut, P, Leroy, V, Gay, F, Lefrancois, D, Bretin, F, Marquette, J.P, Michot, N, Caron, A, Castell, C, Schio, L, McCort, G, Goulaouic, H, Garcia-Echeverria, C, Ronan, B. | | Deposit date: | 2014-08-12 | | Release date: | 2014-11-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Discovery of (2S)-8-[(3R)-3-Methylmorpholin-4-Yl]-1-(3-Methyl-2-Oxobutyl)-2-(Trifluoromethyl)-3,4-Dihydro-2H-Pyrimido[1,2-A]Pyrimidin-6-One: A Novel Potent and Selective Inhibitor of Vps34 for the Treatment of Solid Tumors.
J.Med.Chem., 58, 2015
|
|
7ADW
 
 | | Structure of SARS-CoV-2 Main Protease bound to 2,4'-Dimethylpropiophenone. | | Descriptor: | 2-methyl-1-(4-methylphenyl)propan-1-one, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-09-16 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7A1U
 
 | | Structure of SARS-CoV-2 Main Protease bound to Fusidic Acid. | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, FUSIDIC ACID, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Andaleeb, H, Werner, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-08-14 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
4WA1
 
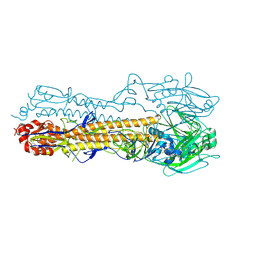 | | The crystal structure of hemagglutinin from a H3N8 influenza virus isolated from New England harbor seals | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Yang, H, Villanueva, J.M, Gubareva, L.V, Stevens, J. | | Deposit date: | 2014-08-28 | | Release date: | 2015-01-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Structural and Functional Analysis of Surface Proteins from an A(H3N8) Influenza Virus Isolated from New England Harbor Seals.
J.Virol., 89, 2015
|
|
4UX5
 
 | | Structure of DNA complex of PCG2 | | Descriptor: | 5'-D(*CP*AP*AP*TP*GP*AP*CP*GP*CP*GP*TP*AP*AP*GP)-3', 5'-D(*CP*TP*TP*AP*CP*GP*CP*GP*TP*CP*AP*TP*TP*GP)-3', TRANSCRIPTION FACTOR MBP1 | | Authors: | Liu, J, Huang, J, Zhao, Y, Liu, H, Wang, D, Yang, J, Zhao, W, Taylor, I.A, Peng, Y. | | Deposit date: | 2014-08-19 | | Release date: | 2015-01-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of DNA Recognition by Pcg2 Reveals a Novel DNA Binding Mode for Winged Helix-Turn-Helix Domains.
Nucleic Acids Res., 43, 2015
|
|
4WA3
 
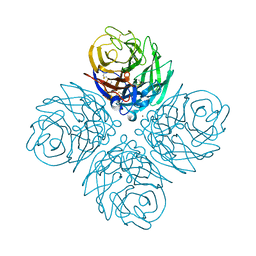 | | The crystal structure of neuraminidase from a H3N8 influenza virus isolated from New England harbor seals | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NICKEL (II) ION, ... | | Authors: | Yang, H, Villanueva, J.M, Gubareva, L.V, Stevens, J. | | Deposit date: | 2014-08-28 | | Release date: | 2015-01-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structural and Functional Analysis of Surface Proteins from an A(H3N8) Influenza Virus Isolated from New England Harbor Seals.
J.Virol., 89, 2015
|
|
4V7J
 
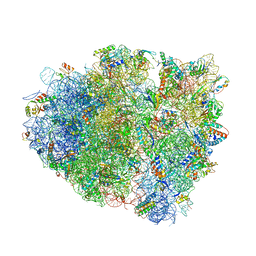 | | Structure of RelE nuclease bound to the 70S ribosome (precleavage state) | | Descriptor: | 30S ribosomal protein S10, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Neubauer, C, Gao, Y.-G, Andersen, K.R, Dunham, C.M, Kelley, A.C, Hentschel, J, Gerdes, K, Ramakrishnan, V, Brodersen, D.E. | | Deposit date: | 2009-11-02 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The structural basis for mRNA recognition and cleavage by the ribosome-dependent endonuclease RelE.
Cell(Cambridge,Mass.), 139, 2009
|
|
4V84
 
 | | Crystal structure of a complex containing domain 3 of CrPV IGR IRES RNA bound to the 70S ribosome. | | Descriptor: | 23S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Zhu, J, Korostelev, A, Costantino, D, Noller, H.F, Kieft, J.S. | | Deposit date: | 2010-12-13 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structures of complexes containing domains from two viral internal ribosome entry site (IRES) RNAs bound to the 70S ribosome.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4V9M
 
 | | 70S Ribosome translocation intermediate FA-4.2A containing elongation factor EFG/FUSIDIC ACID/GDP, mRNA, and tRNA bound in the pe*/E state. | | Descriptor: | 23S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Zhou, J, Lancaster, L, Donohue, J.P, Noller, H.F. | | Deposit date: | 2013-04-25 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Crystal structures of EF-G-ribosome complexes trapped in intermediate states of translocation.
Science, 340, 2013
|
|
4W60
 
 | | The structure of Vaccina virus H7 protein displays A Novel Phosphoinositide binding fold required for membrane biogenesis | | Descriptor: | Late protein H7 | | Authors: | Kolli, S, Meng, X, Wu, X, Shengjuler, D, Cameron, C.E, Xiang, Y, Deng, J. | | Deposit date: | 2014-08-19 | | Release date: | 2014-12-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-function analysis of vaccinia virus h7 protein reveals a novel phosphoinositide binding fold essential for poxvirus replication.
J.Virol., 89, 2015
|
|
4W8E
 
 | | Structure of MST3 with a pyrrolopyrimidine inhibitor (PF-06645342) | | Descriptor: | 3-{4-[(2R)-2-(5-methyl-1,2,4-oxadiazol-3-yl)morpholin-4-yl]-7H-pyrrolo[2,3-d]pyrimidin-5-yl}benzonitrile, Serine/threonine-protein kinase 24 36 kDa subunit | | Authors: | Jasti, J, Song, X, Griffor, M, Kurumbail, R.G. | | Deposit date: | 2014-08-24 | | Release date: | 2015-03-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Discovery and preclinical profiling of 3-[4-(morpholin-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-5-yl]benzonitrile (PF-06447475), a highly potent, selective, brain penetrant, and in vivo active LRRK2 kinase inhibitor.
J.Med.Chem., 58, 2015
|
|
4URL
 
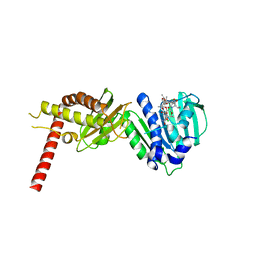 | | Crystal Structure of Staph ParE43kDa in complex with KBD | | Descriptor: | (1R,4aS,5S,6S,8aR)-5-{[(5S)-1-(3-O-acetyl-4-O-carbamoyl-6-deoxy-2-O-methyl-alpha-L-talopyranosyl)-4-hydroxy-2-oxo-5-(propan-2-yl)-2,5-dihydro-1H-pyrrol-3-yl]carbonyl}-6-methyl-4-methylidene-1,2,3,4,4a,5,6,8a-octahydronaphthalen-1-yl 2,6-dideoxy-3-C-[(1S)-1-{[(3,4-dichloro-5-methyl-1H-pyrrol-2-yl)carbonyl]amino}ethyl]-beta-D-ribo-hexopyranoside, DNA TOPOISOMERASE IV, B SUBUNIT | | Authors: | Lu, J, Patel, S, Sharma, N, Soisson, S, Kishii, R, Takei, M, Fukuda, Y, Lumb, K.J, Singh, S.B. | | Deposit date: | 2014-06-30 | | Release date: | 2014-07-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structures of Kibdelomycin Bound to Staphylococcus Aureus Gyrb and Pare Showed a Novel U-Shaped Binding Mode.
Acs Chem.Biol., 9, 2014
|
|
4URT
 
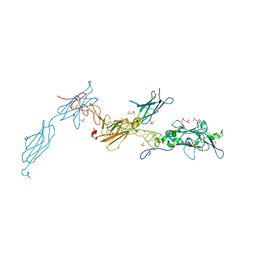 | | The crystal structure of a fragment of netrin-1 in complex with FN5- FN6 of DCC | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Finci, L.I, Krueger, N, Sun, X, Zhang, J, Chegkazi, M, Wu, Y, Schenk, G, Mertens, H.D.T, Svergun, D.I, Zhang, Y, Wang, J.-h, Meijers, R. | | Deposit date: | 2014-07-02 | | Release date: | 2014-09-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The Crystal Structure of Netrin-1 in Complex with Dcc Reveals the Bi-Functionality of Netrin-1 as a Guidance Cue
Neuron, 83, 2014
|
|
4UU3
 
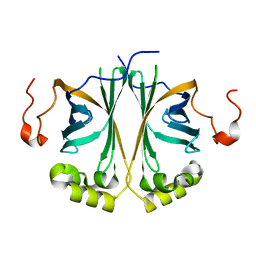 | | Ferulic acid decarboxylase from Enterobacter sp. | | Descriptor: | FERULIC ACID DECARBOXYLASE | | Authors: | Hromic, A, Pavkov-Keller, T, Steinkellner, G, Lyskowski, A, Wuensch, C, Gross, J, Fuchs, M, Fauland, K, Glueck, S.M, Faber, K, Gruber, K. | | Deposit date: | 2014-07-24 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Regioselective Enzymatic Beta-Carboxylation of Para-Hydroxy-Styrene Derivatives Catalyzed by Phenolic Acid Decarboxylases.
Adv. Synth. Catal., 357, 2015
|
|
4UXS
 
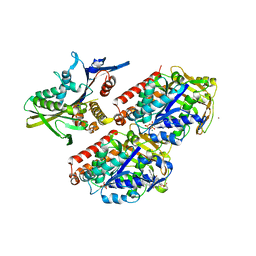 | | Conserved mechanisms of microtubule-stimulated ADP release, ATP binding, and force generation in transport kinesins | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, KINESIN-3 MOTOR DOMAIN, ... | | Authors: | Atherton, J, Farabella, I, Yu, I.M, Rosenfeld, S.S, Houdusse, A, Topf, M, Moores, C. | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Conserved Mechanisms of Microtubule-Stimulated Adp Release, ATP Binding, and Force Generation in Transport Kinesins.
Elife, 3, 2014
|
|
4V6K
 
 | | Structural insights into cognate vs. near-cognate discrimination during decoding. | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Agirrezabala, X, Schreiner, E, Trabuco, L.G, Lei, J, Ortiz-Meoz, R.F, Schulten, K, Green, R, Frank, J. | | Deposit date: | 2011-01-07 | | Release date: | 2014-07-09 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (8.25 Å) | | Cite: | Structural insights into cognate versus near-cognate discrimination during decoding.
Embo J., 30, 2011
|
|
4URM
 
 | | Crystal Structure of Staph GyraseB 24kDa in complex with Kibdelomycin | | Descriptor: | (1R,4aS,5S,6S,8aR)-5-{[(5S)-1-(3-O-acetyl-4-O-carbamoyl-6-deoxy-2-O-methyl-alpha-L-talopyranosyl)-4-hydroxy-2-oxo-5-(propan-2-yl)-2,5-dihydro-1H-pyrrol-3-yl]carbonyl}-6-methyl-4-methylidene-1,2,3,4,4a,5,6,8a-octahydronaphthalen-1-yl 2,6-dideoxy-3-C-[(1S)-1-{[(3,4-dichloro-5-methyl-1H-pyrrol-2-yl)carbonyl]amino}ethyl]-beta-D-ribo-hexopyranoside, DNA GYRASE SUBUNIT B | | Authors: | Lu, J, Patel, S, Sharma, N, Soisson, S, Kishii, R, Takei, M, Fukuda, Y, Lumb, K.J, Singh, S.B. | | Deposit date: | 2014-06-30 | | Release date: | 2014-07-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Structures of Kibdelomycin Bound to Staphylococcus Aureus Gyrb and Pare Showed a Novel U-Shaped Binding Mode.
Acs Chem.Biol., 9, 2014
|
|
