3S90
 
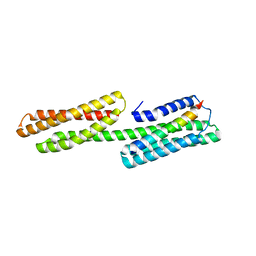 | | Human vinculin head domain Vh1 (residues 1-252) in complex with murine talin (VBS33; residues 1512-1546) | | Descriptor: | Talin-1, Vinculin | | Authors: | Yogesha, S.D, Sharff, A, Bricogne, G, Izard, T. | | Deposit date: | 2011-05-31 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Intermolecular versus intramolecular interactions of the vinculin binding site 33 of talin.
Protein Sci., 20, 2011
|
|
3SMZ
 
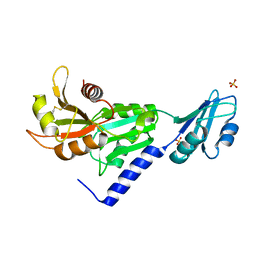 | |
3TJ5
 
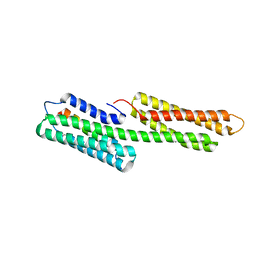 | | human vinculin head domain (Vh1, residues 1-258) in complex with the vinculin binding site of the surface cell antigen 4 (sca4-VBS-N; residues 412-434) from Rickettsia rickettsii | | Descriptor: | Antigenic heat-stable 120 kDa protein, GLYCEROL, Vinculin | | Authors: | Park, H, Lee, J.H, Gouin, E, Cossart, P, Izard, T. | | Deposit date: | 2011-08-23 | | Release date: | 2011-09-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The rickettsia surface cell antigen 4 applies mimicry to bind to and activate vinculin.
J.Biol.Chem., 286, 2011
|
|
3VF0
 
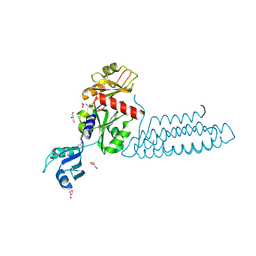 | | Raver1 in complex with metavinculin L954 deletion mutant | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, Ribonucleoprotein PTB-binding 1, ... | | Authors: | Lee, J.H, Vonrhein, C, Bricogne, G, Izard, T. | | Deposit date: | 2012-01-09 | | Release date: | 2012-07-25 | | Last modified: | 2012-09-19 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | The Metavinculin Tail Domain Directs Constitutive Interactions with Raver1 and vinculin RNA.
J.Mol.Biol., 422, 2012
|
|
3U8Z
 
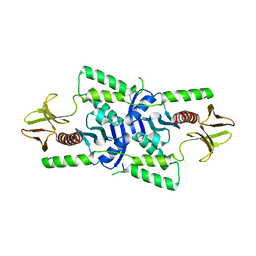 | | human merlin FERM domain | | Descriptor: | Merlin | | Authors: | Yogesha, S.D, Sharff, A.J, Giovannini, M, Bricogne, G, Izard, T. | | Deposit date: | 2011-10-17 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Unfurling of the band 4.1, ezrin, radixin, moesin (FERM) domain of the merlin tumor suppressor.
Protein Sci., 20, 2011
|
|
3TJ6
 
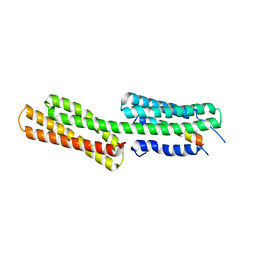 | | human vinculin head domain (Vh1, residues 1-258) in complex with the vinculin binding site of the surface cell antigen 4 (sca4-VBS-C; residues 812-835) from Rickettsia rickettsii | | Descriptor: | Antigenic heat-stable 120 kDa protein, Vinculin | | Authors: | Park, H, Lee, J.H, Gouin, E, Cossart, P, Izard, T. | | Deposit date: | 2011-08-23 | | Release date: | 2011-09-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | The rickettsia surface cell antigen 4 applies mimicry to bind to and activate vinculin.
J.Biol.Chem., 286, 2011
|
|
7RS4
 
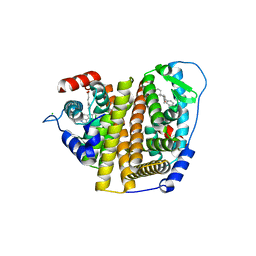 | | Crystal Structure of the ER-alpha Ligand-binding Domain (L372S, L536S) in complex with DMERI-8 | | Descriptor: | (2E)-3-{4-[(1E)-2-(2-chloro-4-fluorophenyl)-1-(2H-indazol-5-yl)but-1-en-1-yl]phenyl}prop-2-enoic acid, CHLORIDE ION, Estrogen receptor | | Authors: | Min, J, Nwachukwu, J.C, Min, C.K, Njeri, J.W, Srinivasan, S, Rangarajan, E.S, Nettles, C.C, Yan, S, Houtman, R, Griffin, P.R, Izard, T, Katzenellenbogen, B.S, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2021-08-10 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Dual-mechanism estrogen receptor inhibitors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7RS2
 
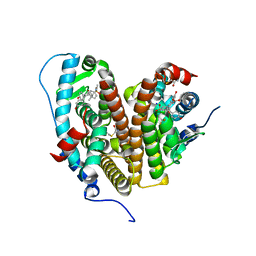 | | Crystal Structure of the ER-alpha Ligand-binding Domain (L372S, L536S) in complex with DMERI-23 | | Descriptor: | (2E)-3-(4-{[(1S,2R,4S,5S,6S)-5,6-bis(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]heptane-2-sulfonyl](2,2,2-trifluoroethyl)amino}phenyl)prop-2-enoic acid, Estrogen receptor | | Authors: | Min, J, Nwachukwu, J.C, Min, C.K, Njeri, J.W, Srinivasan, S, Rangarajan, E.S, Nettles, C.C, Yan, S, Houtman, R, Griffin, P.R, Izard, T, Katzenellenbogen, B.S, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2021-08-10 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Dual-mechanism estrogen receptor inhibitors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7RS9
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (L372S, L536S) in complex with DMERI-25 | | Descriptor: | (1S,2R,4S)-N-[4-(benzyloxy)phenyl]-5,6-bis(4-hydroxyphenyl)-N-(2,2,2-trifluoroethyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, Estrogen receptor | | Authors: | Min, J, Nwachukwu, J.C, Min, C.K, Njeri, J.W, Srinivasan, S, Rangarajan, E.S, Nettles, C.C, Yan, S, Houtman, R, Griffin, P.R, Izard, T, Katzenellenbogen, B.S, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2021-08-11 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Dual-mechanism estrogen receptor inhibitors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7RS1
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (L372S, L536S) in complex with DMERI-21 | | Descriptor: | Estrogen receptor, methyl 3-(4-{[(1S,2R,4S)-5,6-bis(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonyl](2,2,2-trifluoroethyl)amino}phenyl)prop-2-enoate | | Authors: | Min, J, Nwachukwu, J.C, Min, C.K, Njeri, J.W, Srinivasan, S, Rangarajan, E.S, Nettles, C.C, Yan, S, Houtman, R, Griffin, P.R, Izard, T, Katzenellenbogen, B.S, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2021-08-10 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Dual-mechanism estrogen receptor inhibitors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7RS3
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (L372S, L536S) in complex with DMERI-29 | | Descriptor: | (1S,2R,4S)-6-[4-(benzyloxy)phenyl]-5-(4-hydroxyphenyl)-N-(4-methoxyphenyl)-N-(2,2,2-trifluoroethyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, CYSTEINE, Estrogen receptor, ... | | Authors: | Min, J, Nwachukwu, J.C, Min, C.K, Njeri, J.W, Srinivasan, S, Rangarajan, E.S, Nettles, C.C, Yan, S, Houtman, R, Griffin, P.R, Izard, T, Katzenellenbogen, B.S, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2021-08-10 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Dual-mechanism estrogen receptor inhibitors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7RRY
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (L372S, L536S) in complex with DMERI-20 | | Descriptor: | (1S,2R,4S,5S,6S)-5,6-bis(4-hydroxyphenyl)-N-{4-[3-(piperidin-1-yl)propoxy]phenyl}-N-(2,2,2-trifluoroethyl)-7-oxabicyclo[2.2.1]heptane-2-sulfonamide, CHLORIDE ION, Estrogen receptor | | Authors: | Min, J, Nwachukwu, J.C, Min, C.K, Njeri, J.W, Srinivasan, S, Rangarajan, E.S, Nettles, C.C, Yan, S, Houtman, R, Griffin, P.R, Izard, T, Katzenellenbogen, B.S, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2021-08-10 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Dual-mechanism estrogen receptor inhibitors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7RRZ
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (L372S, L536S) in complex with DMERI-30 | | Descriptor: | (1S,2R,4S,5R,6S)-5-(4-hydroxyphenyl)-N-(4-methoxyphenyl)-6-{4-[3-(piperidin-1-yl)propoxy]phenyl}-N-(2,2,2-trifluoroethyl)-7-oxabicyclo[2.2.1]heptane-2-sulfonamide, Estrogen receptor | | Authors: | Min, J, Nwachukwu, J.C, Min, C.K, Njeri, J.W, Srinivasan, S, Rangarajan, E.S, Nettles, C.C, Yan, S, Houtman, R, Griffin, P.R, Izard, T, Katzenellenbogen, B.S, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2021-08-10 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Dual-mechanism estrogen receptor inhibitors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7RS0
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (L372S, L536S) in complex with DMERI-18 | | Descriptor: | (1R,2S,4R,5R,6R)-5-(4-hydroxyphenyl)-N-(4-methoxyphenyl)-6-(4-propoxyphenyl)-N-(2,2,2-trifluoroethyl)-7-oxabicyclo[2.2.1]heptane-2-sulfonamide, Estrogen receptor | | Authors: | Min, J, Nwachukwu, J.C, Min, C.K, Njeri, J.W, Srinivasan, S, Rangarajan, E.S, Nettles, C.C, Yan, S, Houtman, R, Griffin, P.R, Izard, T, Katzenellenbogen, B.S, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2021-08-10 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Dual-mechanism estrogen receptor inhibitors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7RS8
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (L372S, L536S) in complex with DMERI-16 | | Descriptor: | (1R,2S,4R)-5-(4-hydroxyphenyl)-N-(4-methoxyphenyl)-6-{4-[2-(piperidin-1-yl)ethoxy]phenyl}-N-(2,2,2-trifluoroethyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, Estrogen receptor | | Authors: | Min, J, Nwachukwu, J.C, Min, C.K, Njeri, J.W, Srinivasan, S, Rangarajan, E.S, Nettles, C.C, Yan, S, Houtman, R, Griffin, P.R, Izard, T, Katzenellenbogen, B.S, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2021-08-11 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Dual-mechanism estrogen receptor inhibitors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7RRX
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (L372S, L536S) in complex with DMERI-19 | | Descriptor: | (1S,2R,4S)-5,6-bis(4-hydroxyphenyl)-N-{4-[2-(piperidin-1-yl)ethoxy]phenyl}-N-(2,2,2-trifluoroethyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, CHLORIDE ION, Estrogen receptor | | Authors: | Min, J, Nwachukwu, J.C, Min, C.K, Njeri, J.W, Srinivasan, S, Rangarajan, E.S, Nettles, C.C, Yan, S, Houtman, R, Griffin, P.R, Izard, T, Katzenellenbogen, B.S, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2021-08-10 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Dual-mechanism estrogen receptor inhibitors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7RS7
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (L372S, L536S) in complex with DMERI-30 | | Descriptor: | (1S,2R,4S,5S,6S)-N,5,6-tris(4-hydroxyphenyl)-N-(2,2,2-trifluoroethyl)-7-oxabicyclo[2.2.1]heptane-2-sulfonamide, Estrogen receptor | | Authors: | Min, J, Nwachukwu, J.C, Min, C.K, Njeri, J.W, Srinivasan, S, Rangarajan, E.S, Nettles, C.C, Yan, S, Houtman, R, Griffin, P.R, Izard, T, Katzenellenbogen, B.S, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2021-08-11 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Dual-mechanism estrogen receptor inhibitors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5L0C
 
 | | Human metavinculin (residues 959-1134) in complex with PIP2 | | Descriptor: | PHOSPHATE ION, Vinculin, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Chinthalapudi, K, Izard, T. | | Deposit date: | 2016-07-27 | | Release date: | 2016-08-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Differential lipid binding of vinculin isoforms promotes quasi-equivalent dimerization.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5L0F
 
 | |
5L0G
 
 | | Human metavinculin MVt Q971R, R975D, T978R mutant (residues 959-1134) in complex with PIP2 | | Descriptor: | Vinculin, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Chinthalapudi, K, Izard, T. | | Deposit date: | 2016-07-27 | | Release date: | 2016-08-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Differential lipid binding of vinculin isoforms promotes quasi-equivalent dimerization.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5L0D
 
 | | Human Metavinculin(residues 959-1130) in complex with PIP2 | | Descriptor: | Vinculin, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Chinthalapudi, K, Izard, T. | | Deposit date: | 2016-07-27 | | Release date: | 2016-08-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Differential lipid binding of vinculin isoforms promotes quasi-equivalent dimerization.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5L0H
 
 | |
5L0I
 
 | |
5L0J
 
 | |
1TFU
 
 | |
