3L6Y
 
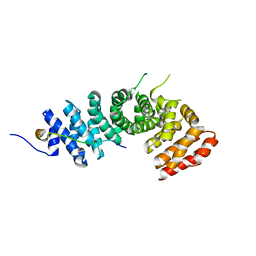 | | Crystal structure of p120 catenin in complex with E-cadherin | | Descriptor: | Catenin delta-1, E-cadherin | | Authors: | Ishiyama, N, Lee, S.-H, Liu, S, Li, G.-Y, Smith, M.J, Reichardt, L.F, Ikura, M. | | Deposit date: | 2009-12-27 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Dynamic and static interactions between p120 catenin and E-cadherin regulate the stability of cell-cell adhesion.
Cell(Cambridge,Mass.), 141, 2010
|
|
3L6X
 
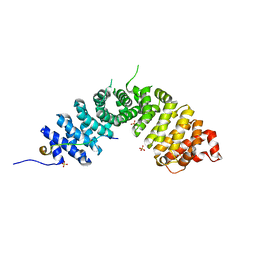 | | Crystal structure of p120 catenin in complex with E-cadherin | | Descriptor: | Catenin delta-1, E-cadherin, SULFATE ION | | Authors: | Ishiyama, N, Lee, S.-H, Liu, S, Li, G.-Y, Smith, M.J, Reichardt, L.F, Ikura, M. | | Deposit date: | 2009-12-27 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Dynamic and static interactions between p120 catenin and E-cadherin regulate the stability of cell-cell adhesion.
Cell(Cambridge,Mass.), 141, 2010
|
|
4K1N
 
 | |
4K1O
 
 | |
6DUW
 
 | |
6DV1
 
 | |
6DUY
 
 | |
1SB9
 
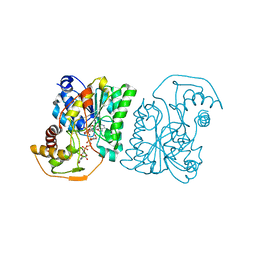 | | Crystal structure of Pseudomonas aeruginosa UDP-N-acetylglucosamine 4-epimerase complexed with UDP-glucose | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, URIDINE-5'-DIPHOSPHATE-GLUCOSE, wbpP | | Authors: | Ishiyama, N, Creuzenet, C, Lam, J.S, Berghuis, A.M. | | Deposit date: | 2004-02-10 | | Release date: | 2004-05-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of WbpP, a Genuine UDP-N-acetylglucosamine 4-Epimerase from Pseudomonas aeruginosa: SUBSTRATE SPECIFICITY IN UDP-HEXOSE 4-EPIMERASES.
J.Biol.Chem., 279, 2004
|
|
1SB8
 
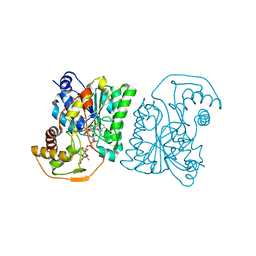 | | Crystal structure of Pseudomonas aeruginosa UDP-N-acetylglucosamine 4-epimerase complexed with UDP-N-acetylgalactosamine | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, URIDINE-DIPHOSPHATE-N-ACETYLGALACTOSAMINE, wbpP | | Authors: | Ishiyama, N, Creuzenet, C, Lam, J.S, Berghuis, A.M. | | Deposit date: | 2004-02-10 | | Release date: | 2004-05-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of WbpP, a Genuine UDP-N-acetylglucosamine 4-Epimerase from Pseudomonas aeruginosa: SUBSTRATE SPECIFICITY IN UDP-HEXOSE 4-EPIMERASES.
J.Biol.Chem., 279, 2004
|
|
2GNA
 
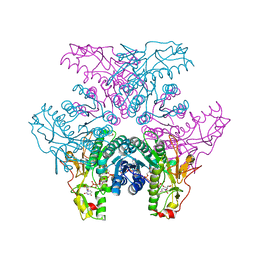 | | Crystal structure of UDP-GlcNAc inverting 4,6-dehydratase in complex with NADP and UDP-Gal | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GALACTOSE-URIDINE-5'-DIPHOSPHATE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Ishiyama, N, Creuzenet, C, Lam, J.S, Berghuis, A.M. | | Deposit date: | 2006-04-09 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Studies of FlaA1 from Helicobacter pylori Reveal the Mechanism for Inverting 4,6-Dehydratase Activity.
J.Biol.Chem., 281, 2006
|
|
2GN9
 
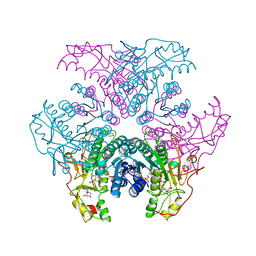 | | Crystal structure of UDP-GlcNAc inverting 4,6-dehydratase in complex with NADP and UDP-Glc | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, UDP-GlcNAc C6 dehydratase, ... | | Authors: | Ishiyama, N, Creuzenet, C, Lam, J.S, Berghuis, A.M. | | Deposit date: | 2006-04-09 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Studies of FlaA1 from Helicobacter pylori Reveal the Mechanism for Inverting 4,6-Dehydratase Activity.
J.Biol.Chem., 281, 2006
|
|
2GN8
 
 | | Crystal structure of UDP-GlcNAc inverting 4,6-dehydratase in complex with NADP and UDP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, UDP-GlcNAc C6 dehydratase, ... | | Authors: | Ishiyama, N, Creuzenet, C, Lam, J.S, Berghuis, A.M. | | Deposit date: | 2006-04-09 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Studies of FlaA1 from Helicobacter pylori Reveal the Mechanism for Inverting 4,6-Dehydratase Activity.
J.Biol.Chem., 281, 2006
|
|
2GN6
 
 | | Crystal structure of UDP-GlcNAc inverting 4,6-dehydratase in complex with NADP and UDP-GlcNAc | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, UDP-GlcNAc C6 dehydratase, ... | | Authors: | Ishiyama, N, Creuzenet, C, Lam, J.S, Berghuis, A.M. | | Deposit date: | 2006-04-09 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Studies of FlaA1 from Helicobacter pylori Reveal the Mechanism for Inverting 4,6-Dehydratase Activity.
J.Biol.Chem., 281, 2006
|
|
2GN4
 
 | | Crystal structure of UDP-GlcNAc inverting 4,6-dehydratase in complex with NADPH and UDP-GlcNAc | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, UDP-GlcNAc C6 dehydratase, ... | | Authors: | Ishiyama, N, Creuzenet, C, Lam, J.S, Berghuis, A.M. | | Deposit date: | 2006-04-09 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Studies of FlaA1 from Helicobacter pylori Reveal the Mechanism for Inverting 4,6-Dehydratase Activity.
J.Biol.Chem., 281, 2006
|
|
3JRR
 
 | | Crystal structure of the ligand binding suppressor domain of type 3 inositol 1,4,5-trisphosphate receptor | | Descriptor: | Inositol 1,4,5-trisphosphate receptor type 3 | | Authors: | Chan, J, Ishiyama, N, Ikura, M. | | Deposit date: | 2009-09-08 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A 1.9 angstrom crystal structure of the suppressor domain of type 3 inositol 1,4,5-trisphosphate receptor
To be Published
|
|
5WI4
 
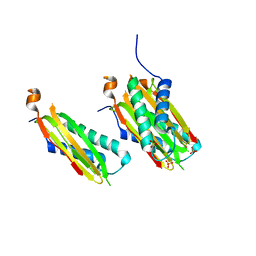 | | CRYSTAL STRUCTURE OF DYNLT1/TCTEX-1 IN COMPLEX WITH ARHGEF2 | | Descriptor: | Dynein light chain Tctex-type 1,Rho guanine nucleotide exchange factor 2, SULFATE ION | | Authors: | Balan, M, Ishiyama, N, Marshall, C.B, Ikura, M. | | Deposit date: | 2017-07-18 | | Release date: | 2017-11-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | MARK3-mediated phosphorylation of ARHGEF2 couples microtubules to the actin cytoskeleton to establish cell polarity.
Sci Signal, 10, 2017
|
|
6PW7
 
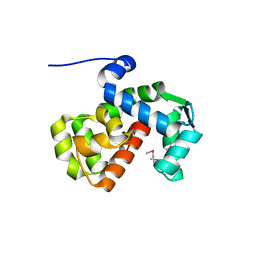 | | X-ray crystal structure of C. elegans STIM EF-SAM domain | | Descriptor: | CALCIUM ION, Stromal interaction molecule 1 | | Authors: | Enomoto, M, Nishikawa, T, Back, S.I, Ishiyama, N, Zheng, L, Stathopulos, P.B, Ikura, M. | | Deposit date: | 2019-07-22 | | Release date: | 2019-11-13 | | Last modified: | 2020-02-12 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Coordination of a Single Calcium Ion in the EF-hand Maintains the Off State of the Stromal Interaction Molecule Luminal Domain.
J.Mol.Biol., 432, 2020
|
|
6OS4
 
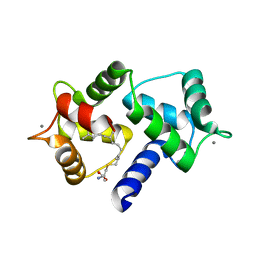 | | Calmodulin in complex with farnesyl cysteine methyl ester | | Descriptor: | CALCIUM ION, Calmodulin-1, s-farnesyl-l-cysteine methyl ester | | Authors: | Grant, B.M.M, Enomoto, M, Lee, K.Y, Back, S.I, Gebregiworgis, T, Ishiyama, N, Ikura, M, Marshall, C. | | Deposit date: | 2019-05-01 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Calmodulin disrupts plasma membrane localization of farnesylated KRAS4b by sequestering its lipid moiety.
Sci.Signal., 13, 2020
|
|
3HSM
 
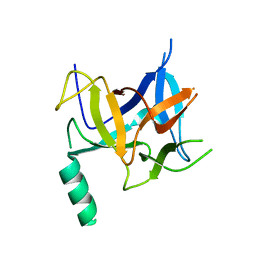 | | Crystal structure of distal N-terminal beta-trefoil domain of Ryanodine Receptor type 1 | | Descriptor: | Ryanodine receptor 1 | | Authors: | Amador, F.J, Liu, S, Ishiyama, N, Plevin, M.J, Wilson, A, MacLennan, D.H, Ikura, M. | | Deposit date: | 2009-06-10 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of type I ryanodine receptor amino-terminal beta-trefoil domain reveals a disease-associated mutation "hot spot" loop
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3SEA
 
 | | Structure of Rheb-Y35A mutant in GDP- and GMPPNP-bound forms | | Descriptor: | ACETATE ION, GTP-binding protein Rheb, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Mazhab-Jafari, M.T, Marshall, C.B, Ishiyama, N, Vuk, S, Ikura, M. | | Deposit date: | 2011-06-10 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An Autoinhibited Noncanonical Mechanism of GTP Hydrolysis by Rheb Maintains mTORC1 Homeostasis.
Structure, 20, 2012
|
|
4O2L
 
 | | Structure of Mus musculus Rheb G63A mutant bound to GTP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, GTP-binding protein Rheb, ... | | Authors: | Mazhab-Jafari, M.T, Marshall, C.B, Ho, J, Ishiyama, N, Stambolic, V, Ikura, M. | | Deposit date: | 2013-12-17 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-guided mutation of the conserved G3-box glycine in Rheb generates a constitutively activated regulator of mammalian target of rapamycin (mTOR).
J.Biol.Chem., 289, 2014
|
|
4O2R
 
 | | Structure of Mus musculus Rheb G63V mutant bound to GDP | | Descriptor: | GTP-binding protein Rheb, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Mazhab-Jafari, M.T, Marshall, C.B, Ho, J, Ishiyama, N, Stambolic, V, Ikura, M. | | Deposit date: | 2013-12-17 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-guided mutation of the conserved G3-box glycine in Rheb generates a constitutively activated regulator of mammalian target of rapamycin (mTOR).
J.Biol.Chem., 289, 2014
|
|
4O25
 
 | | Structure of Wild Type Mus musculus Rheb bound to GTP | | Descriptor: | GTP-binding protein Rheb, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Mazhab-Jafari, M.T, Marshall, C.B, Ho, J, Ishiyama, N, Stambolic, V, Ikura, M. | | Deposit date: | 2013-12-16 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-guided mutation of the conserved G3-box glycine in Rheb generates a constitutively activated regulator of mammalian target of rapamycin (mTOR).
J.Biol.Chem., 289, 2014
|
|
6AMB
 
 | | Crystal Structure of the Afadin RA1 domain in complex with HRAS | | Descriptor: | Afadin, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Smith, M.J, Ishiyama, N, Ikura, M. | | Deposit date: | 2017-08-09 | | Release date: | 2017-11-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Evolution of AF6-RAS association and its implications in mixed-lineage leukemia.
Nat Commun, 8, 2017
|
|
3UJ4
 
 | | Crystal structure of the apo-inositol 1,4,5-trisphosphate receptor | | Descriptor: | Inositol 1,4,5-trisphosphate receptor type 1, SULFATE ION | | Authors: | Ikura, M, Seo, M.D, Ishiyama, N, Stathopulos, P. | | Deposit date: | 2011-11-07 | | Release date: | 2012-02-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and functional conservation of key domains in InsP3 and ryanodine receptors.
Nature, 483, 2012
|
|
