8EON
 
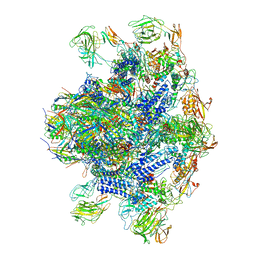 | | Pseudomonas phage E217 baseplate complex | | Descriptor: | Baseplate component gp33, Baseplate component gp34, Baseplate component gp36, ... | | Authors: | Li, F, Cingolani, G, Hou, C. | | Deposit date: | 2022-10-03 | | Release date: | 2023-07-26 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | High-resolution cryo-EM structure of the Pseudomonas bacteriophage E217.
Nat Commun, 14, 2023
|
|
8ENV
 
 | | In situ cryo-EM structure of Pseudomonas phage E217 tail baseplate in C6 map | | Descriptor: | Baseplate_J domain-containing protein gp44, Ripcord gp36, Sheath initiator gp34, ... | | Authors: | Li, F, Cingolani, G, Hou, C. | | Deposit date: | 2022-09-30 | | Release date: | 2023-07-26 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | High-resolution cryo-EM structure of the Pseudomonas bacteriophage E217.
Nat Commun, 14, 2023
|
|
8FRS
 
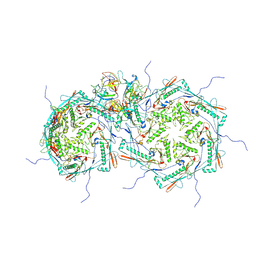 | |
8FVH
 
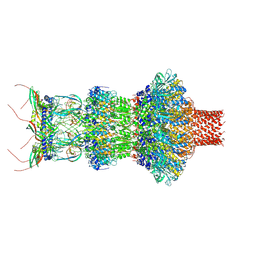 | | Pseudomonas phage E217 neck (portal, head-to-tail connector, collar and gateway proteins) | | Descriptor: | E217 collar protein gp28, E217 gateway protein gp29, E217 head-to-tail connector protein gp27, ... | | Authors: | Li, F, Cingolani, G, Hou, C. | | Deposit date: | 2023-01-18 | | Release date: | 2023-07-26 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | High-resolution cryo-EM structure of the Pseudomonas bacteriophage E217.
Nat Commun, 14, 2023
|
|
6Z47
 
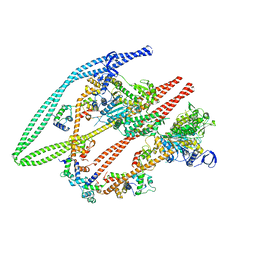 | | Smooth muscle myosin shutdown state heads region | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Myosin heavy chain 11, ... | | Authors: | Scarff, C.A, Carrington, G, Casas Mao, D, Chalovich, J.M, Knight, P.J, Ranson, N.A, Peckham, M. | | Deposit date: | 2020-05-22 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | Structure of the shutdown state of myosin-2.
Nature, 588, 2020
|
|
5GZO
 
 | | Structure of neutralizing antibody bound to Zika envelope protein | | Descriptor: | Antibody heavy chain, Antibody light chain, Genome polyprotein | | Authors: | Wang, Q, Yang, H, Liu, X, Dai, L, Ma, T, Qi, J, Wong, G, Peng, R, Liu, S, Li, J, Li, S, Song, J, Liu, J, He, J, Yuan, H, Xiong, Y, Liao, Y, Li, J, Yang, J, Tong, Z, Griffin, B, Bi, Y, Liang, M, Xu, X, Cheng, G, Wang, P, Qiu, X, Kobinger, G, Shi, Y, Yan, J, Gao, G.F. | | Deposit date: | 2016-09-29 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.755 Å) | | Cite: | Molecular determinants of human neutralizing antibodies isolated from a patient infected with Zika virus
Sci Transl Med, 8, 2016
|
|
4PP7
 
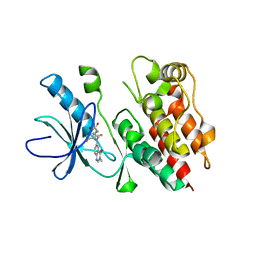 | | Highly Potent and Selective 3-N-methylquinazoline-4(3H)-one Based Inhibitors of B-RafV600E Kinase | | Descriptor: | N-{2,4-difluoro-3-[methyl(3-methyl-4-oxo-3,4-dihydroquinazolin-6-yl)amino]phenyl}propane-1-sulfonamide, Serine/threonine-protein kinase B-raf | | Authors: | Wenglowsky, S, Ren, L, Grina, J, Hansen, J.D, Laird, E.R, Moreno, D, Dinkel, V, Gloor, S.L, Hastings, G, Rana, S, Rasor, K, Sturgis, H.L, Voegtli, W.C, Vigers, G.P.A, Willis, B, Mathieu, S, Rudolph, J. | | Deposit date: | 2014-02-26 | | Release date: | 2014-04-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Highly potent and selective 3-N-methylquinazoline-4(3H)-one based inhibitors of B-Raf(V600E) kinase.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
1XJI
 
 | | Bacteriorhodopsin crystallized in bicelles at room temperature | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Bacteriorhodopsin, DECANE, ... | | Authors: | Faham, S, Boulting, G.L, Massey, E.A, Yohannan, S, Yang, D, Bowie, J.U. | | Deposit date: | 2004-09-23 | | Release date: | 2005-04-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallization of bacteriorhodopsin from bicelle formulations at room temperature
Protein Sci., 14, 2005
|
|
4PY6
 
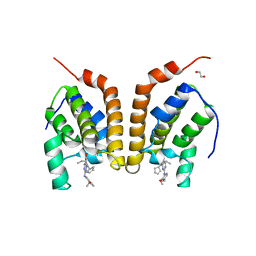 | | Crystal Structure of bromodomain of PFA0510w from Plasmodium Falciparum | | Descriptor: | 1,2-ETHANEDIOL, 4-{[(7R)-8-cyclopentyl-7-ethyl-5-methyl-6-oxo-5,6,7,8-tetrahydropteridin-2-yl]amino}-3-methoxy-N-(1-methylpiperidin-4-yl)benzamide, Bromodomain protein, ... | | Authors: | Fonseca, M, Tallant, C, Hutchinson, A, Savitsky, P, Krojer, T, Filippakopoulos, P, Loppnau, P, Brennan, P.E, von Delft, F, Dong, A, Josling, G.A, Duffy, M.F, Arrowsmith, C.H, Bountra, C, Hui, R, Knapp, S, Wernimont, A.K, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-03-26 | | Release date: | 2014-04-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of bromodomain of PFA0510w from Plasmodium Falciparum
To be Published
|
|
8TVU
 
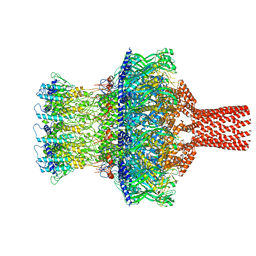 | |
8TVR
 
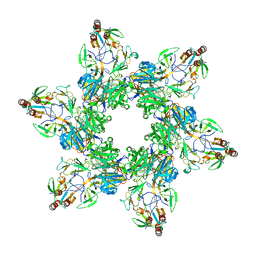 | |
1Y2A
 
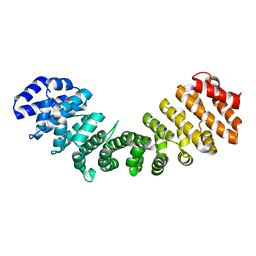 | | Structure of mammalian importin bound to the non-classical PLSCR1-NLS | | Descriptor: | Importin alpha-2 Subunit, decamer fragment of Phospholipid scramblase 1 | | Authors: | Chen, M.-H, Ben-Efraim, I, Mitrousis, G, Walker-Kopp, N, Sims, P.J, Cingolani, G. | | Deposit date: | 2004-11-22 | | Release date: | 2005-02-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Phospholipid Scramblase 1 Contains a Nonclassical Nuclear Localization Signal with Unique Binding Site in Importin alpha
J.Biol.Chem., 280, 2005
|
|
6V6T
 
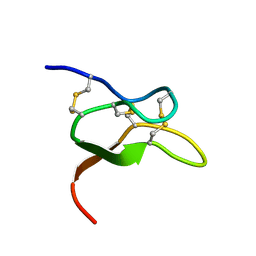 | |
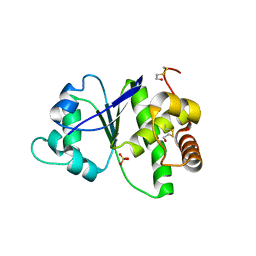
4R30
 
 | |
8U10
 
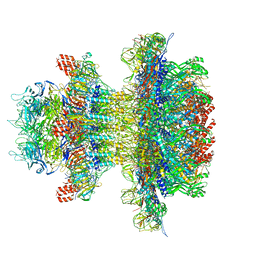 | | In situ cryo-EM structure of bacteriophage P22 gp1:gp4:gp5:gp10:gp9 N-term complex in conformation 1 at 3.2A resolution | | Descriptor: | Major capsid protein, Packaged DNA stabilization protein gp10, Peptidoglycan hydrolase gp4, ... | | Authors: | Iglesias, S, Feng-Hou, C, Cingolani, G. | | Deposit date: | 2023-08-30 | | Release date: | 2023-11-22 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular Architecture of Salmonella Typhimurium Virus P22 Genome Ejection Machinery.
J.Mol.Biol., 435, 2023
|
|
8U11
 
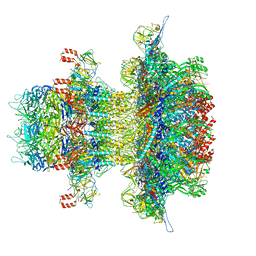 | | In situ cryo-EM structure of bacteriophage P22 gp1:gp5:gp4: gp10: gp9 N-term complex in conformation 2 at 3.1A resolution | | Descriptor: | Major capsid protein, Packaged DNA stabilization protein gp10, Peptidoglycan hydrolase gp4, ... | | Authors: | Iglesias, S, Feng-Hou, C, Cingolani, G. | | Deposit date: | 2023-08-30 | | Release date: | 2023-11-22 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular Architecture of Salmonella Typhimurium Virus P22 Genome Ejection Machinery.
J.Mol.Biol., 435, 2023
|
|
4TTO
 
 | |
4TTM
 
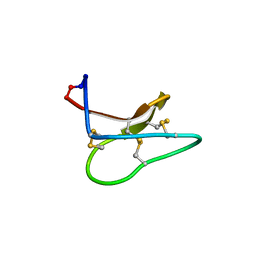 | | Racemic structure of kalata B1 (kB1) | | Descriptor: | D-kalata B1, Kalata-B1 | | Authors: | Wang, C.K, King, G.J, Craik, D.J. | | Deposit date: | 2014-06-22 | | Release date: | 2014-09-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9001 Å) | | Cite: | Racemic and Quasi-Racemic X-ray Structures of Cyclic Disulfide-Rich Peptide Drug Scaffolds.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4TTL
 
 | |
6C54
 
 | | Ebola nucleoprotein nucleocapsid-like assembly and the asymmetric unit | | Descriptor: | Nucleoprotein | | Authors: | Su, Z, Wu, C, Pintilie, G.D, Chiu, W, Amarasinghe, G.K, Leung, D.W. | | Deposit date: | 2018-01-13 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Electron Cryo-microscopy Structure of Ebola Virus Nucleoprotein Reveals a Mechanism for Nucleocapsid-like Assembly.
Cell, 172, 2018
|
|
6WI6
 
 | | Crystal structure of plantacyclin B21AG | | Descriptor: | MALONATE ION, Plantacyclin B21AG | | Authors: | Smith, A.T, Gor, M.C, Vezina, B, McMahon, R, King, G, Panjikar, S, Rehm, B, Martin, J. | | Deposit date: | 2020-04-08 | | Release date: | 2021-01-06 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure and site-directed mutagenesis of circular bacteriocin plantacyclin B21AG reveals cationic and aromatic residues important for antimicrobial activity.
Sci Rep, 10, 2020
|
|
4GXZ
 
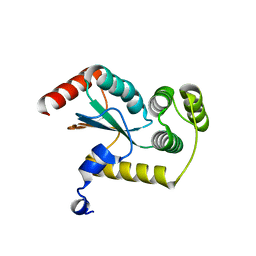 | | Crystal structure of a periplasmic thioredoxin-like protein from Salmonella enterica serovar Typhimurium | | Descriptor: | Suppression of copper sensitivity protein | | Authors: | Shepherd, M, Heras, B, King, G.J, Argente, M.P, Achard, M.E.S, King, N.P, McEwan, A.G, Schembri, M.A. | | Deposit date: | 2012-09-04 | | Release date: | 2013-07-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural and functional characterization of ScsC, a periplasmic thioredoxin-like protein from Salmonella enterica serovar Typhimurium
Antioxid Redox Signal, 19, 2013
|
|
4TTK
 
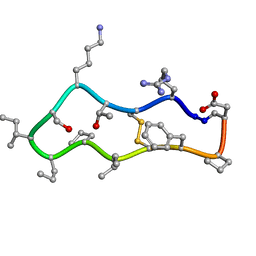 | |
4TTN
 
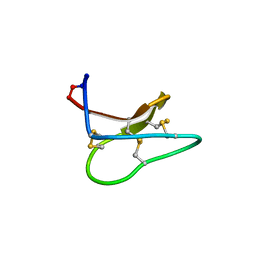 | | Quasi-racemic structure of [G6A]kalata B1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, D-kalata B1, Kalata-B1 | | Authors: | Wang, C.K, King, G.J, Craik, D.J. | | Deposit date: | 2014-06-22 | | Release date: | 2014-09-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.2507 Å) | | Cite: | Racemic and Quasi-Racemic X-ray Structures of Cyclic Disulfide-Rich Peptide Drug Scaffolds.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
1YR1
 
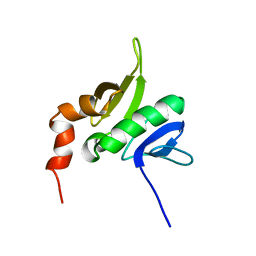 | |
