4WCV
 
 | | Haloalkane dehalogenase DhaA mutant from Rhodococcus rhodochrous (T148L+G171Q+A172V+C176G) | | Descriptor: | ACETATE ION, CHLORIDE ION, Haloalkane dehalogenase, ... | | Authors: | Holubeva, T, Prudnikova, T, Kuta-Smatanova, I, Rezacova, P. | | Deposit date: | 2014-09-05 | | Release date: | 2014-10-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Balancing the stability-activity trade-off by fine-tuning dehalogenase access tunnels
Chemcatchem, 2015
|
|
7AEG
 
 | | SARS-CoV-2 main protease in a covalent complex with SDZ 224015 derivative, compound 5 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-[(benzyloxy)carbonyl]-L-valyl-N-[(1S)-1-(carboxymethyl)-3-fluoro-2-oxopropyl]-L-alaninamide | | Authors: | Owen, C.D, Redhead, M.A, Lukacik, P, Strain-Damerell, C, Fearon, D, Brewitz, L, Collette, A, Robinson, C, Collins, P, Radoux, C, Navratilova, I, Douangamath, A, von Delft, F, Malla, T.R, Nugen, T, Hull, H, Tumber, A, Schofield, C.J, Hallet, D, Stuart, D.I, Hopkins, A.L, Walsh, M.A. | | Deposit date: | 2020-09-17 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Bispecific repurposed medicines targeting the viral and immunological arms of COVID-19.
Sci Rep, 11, 2021
|
|
4UYA
 
 | | Structure of MLK4 kinase domain with ATPgammaS | | Descriptor: | MAGNESIUM ION, MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE MLK4, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Read, J.A, Brassington, C, Pollard, H.K, Phillips, C, Green, I, Overmann, R, Collier, M. | | Deposit date: | 2014-08-29 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Recurrent Mlk4 Loss-of-Function Mutations Suppress Jnk Signaling to Promote Colon Tumorigenesis.
Cancer Res., 76, 2016
|
|
5IY8
 
 | | Human holo-PIC in the initial transcribing state | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB10, DNA-directed RNA polymerase II subunit RPB11-a, ... | | Authors: | He, Y, Yan, C, Fang, J, Inouye, C, Tjian, R, Ivanov, I, Nogales, E. | | Deposit date: | 2016-03-24 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Near-atomic resolution visualization of human transcription promoter opening.
Nature, 533, 2016
|
|
6XC9
 
 | | Immune receptor complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Tran, T.M, Faridi, P, Lim, J.J, Ting, T.Y, Onwukwe, G, Bhattacharjee, P, Tresoldi, M.C.E, Cameron, J.F, La-Gruta, L.N, Purcell, W.A, Mannering, I.S, Rossjohn, J, Reid, H.H. | | Deposit date: | 2020-06-08 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | T cell receptor recognition of hybrid insulin peptides bound to HLA-DQ8.
Nat Commun, 12, 2021
|
|
5IY6
 
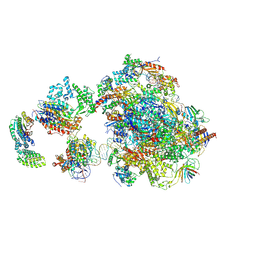 | | Human holo-PIC in the closed state | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB10, DNA-directed RNA polymerase II subunit RPB11-a, ... | | Authors: | He, Y, Yan, C, Fang, J, Inouye, C, Tjian, R, Ivanov, I, Nogales, E. | | Deposit date: | 2016-03-24 | | Release date: | 2016-05-18 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Near-atomic resolution visualization of human transcription promoter opening.
Nature, 533, 2016
|
|
6ZUQ
 
 | | Crystal structure of the effector Ecp11-1 from Fulvia fulva | | Descriptor: | Extracellular protein 11-1, GLYCEROL, ZINC ION | | Authors: | Lazar, N, Mesarich, C, Petit-Houdenot, Y, Talbi, N, Li de la Sierra-Gallay, I, Zelie, E, Blondeau, K, Gracy, J, Ollivier, B, van de Wouw, A, Balesdent, M.H, Idnurm, A, van Tilbeurgh, H, Fudal, I. | | Deposit date: | 2020-07-23 | | Release date: | 2021-08-04 | | Last modified: | 2022-07-27 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | A new family of structurally conserved fungal effectors displays epistatic interactions with plant resistance proteins.
Plos Pathog., 18, 2022
|
|
4UX5
 
 | | Structure of DNA complex of PCG2 | | Descriptor: | 5'-D(*CP*AP*AP*TP*GP*AP*CP*GP*CP*GP*TP*AP*AP*GP)-3', 5'-D(*CP*TP*TP*AP*CP*GP*CP*GP*TP*CP*AP*TP*TP*GP)-3', TRANSCRIPTION FACTOR MBP1 | | Authors: | Liu, J, Huang, J, Zhao, Y, Liu, H, Wang, D, Yang, J, Zhao, W, Taylor, I.A, Peng, Y. | | Deposit date: | 2014-08-19 | | Release date: | 2015-01-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of DNA Recognition by Pcg2 Reveals a Novel DNA Binding Mode for Winged Helix-Turn-Helix Domains.
Nucleic Acids Res., 43, 2015
|
|
4WI7
 
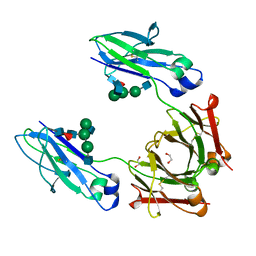 | | Structural mapping of the human IgG1 binding site for FcRn: hu3S193 Fc mutation H435A | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Ig gamma-1 chain C region | | Authors: | Farrugia, W, Burvenich, I.J.G, Scott, A.M, Ramsland, P.A. | | Deposit date: | 2014-09-25 | | Release date: | 2015-09-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional mapping of human IgG1 binding site for FcRn in vivo using human FcRn transgenic mice
To Be Published
|
|
7ABW
 
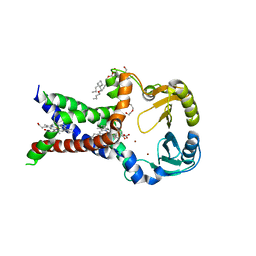 | | Crystal structure of siderophore reductase FoxB | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, DECYL-BETA-D-MALTOPYRANOSIDE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Josts, I, Tidow, H. | | Deposit date: | 2020-09-09 | | Release date: | 2021-09-01 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Structural insights into a novel family of integral membrane siderophore reductases.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4WI8
 
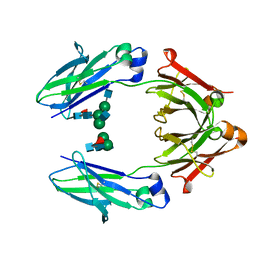 | | Structural mapping of the human IgG1 binding site for FcRn: hu3S193 Fc mutation Y436A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-beta-D-mannopyranose, ... | | Authors: | Farrugia, W, Burvenich, I.J.G, Scott, A.M, Ramsland, P.A. | | Deposit date: | 2014-09-25 | | Release date: | 2015-09-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and functional mapping of human IgG1 binding site for FcRn in vivo using human FcRn transgenic mice
To Be Published
|
|
7AD5
 
 | | Crystal structure of the effector AvrLm5-9 from Leptosphaeria maculans | | Descriptor: | ACETATE ION, Avirulence protein LmJ1, GLYCEROL, ... | | Authors: | Lazar, N, Mesarich, C, Petit-Houdenot, Y, Talbi, N, Li de la Sierra-Gallay, I, Zelie, E, Blondeau, K, Gracy, J, Ollivier, B, van de Wouw, A, Balesdent, M.H, Idnurm, A, van Tilbeurgh, H, Fudal, I. | | Deposit date: | 2020-09-14 | | Release date: | 2021-10-06 | | Last modified: | 2022-07-27 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | A new family of structurally conserved fungal effectors displays epistatic interactions with plant resistance proteins.
Plos Pathog., 18, 2022
|
|
4WH6
 
 | | Crystal structure of HCV NS3/4A protease variant R155K in complex with Asunaprevir | | Descriptor: | Genome polyprotein, N-(tert-butoxycarbonyl)-3-methyl-L-valyl-(4R)-4-[(7-chloro-4-methoxyisoquinolin-1-yl)oxy]-N-{(1R,2S)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethenylcyclopropyl}-L-prolinamide, SULFATE ION, ... | | Authors: | Soumana, D.I, Ali, A, Schiffer, C.A. | | Deposit date: | 2014-09-20 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural Analysis of Asunaprevir Resistance in HCV NS3/4A Protease.
Acs Chem.Biol., 9, 2014
|
|
1B6U
 
 | |
8B2M
 
 | | Matrix-metallopeptidase inhibitor Potempin A (PotA) from Tannerella forsythia | | Descriptor: | NICKEL (II) ION, Tannerella forsythia Potempin A (PotA) | | Authors: | Potempa, J, Ksiazek, M, Goulas, T, Cuppari, A, Rodriguez-Banqueri, A, Arolas, J.L, Lopez-Pelegrin, M, Garcia-Ferrer, I, Guevara, T. | | Deposit date: | 2022-09-14 | | Release date: | 2022-12-21 | | Last modified: | 2023-02-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A unique network of attack, defence and competence on the outer membrane of the periodontitis pathogen Tannerella forsythia.
Chem Sci, 14, 2023
|
|
4WHW
 
 | | Direct photocapture of bromodomains using tropolone chemical probes | | Descriptor: | 1,2-ETHANEDIOL, 2-methoxy-4-{1-[2-(morpholin-4-yl)ethyl]-2-(2-phenylethyl)-1H-benzimidazol-5-yl}cyclohepta-2,4,6-trien-1-one, Bromodomain-containing protein 4 | | Authors: | Hett, E.C, Piatnitski Chekler, E.L, Basak, A, Bonin, P.D, Denny, R.A, Flick, A.C, Geoghegan, K.F, Liu, S, Pletcher, M.T, Robinson, R.P, Sahasrabudhe, P, Salter, S, Stock, I.A, Jones, L.H. | | Deposit date: | 2014-09-23 | | Release date: | 2015-10-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.345 Å) | | Cite: | Direct photocapture of bromodomains using tropolone chemical probes
To Be Published
|
|
4WPG
 
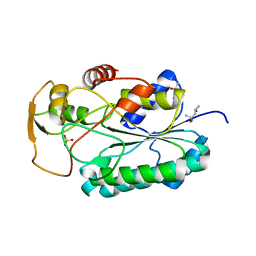 | | Group A Streptococcus GacA is an essential dTDP-4-dehydrorhamnose reductase (RmlD) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, PHOSPHATE ION, ... | | Authors: | van der Beek, S.L, Le Breton, Y, Ferenbach, A.T, van Aalten, D.M.F, Navratilova, I, McIver, K, van Sorge, N.M, Dorfmueller, H.C. | | Deposit date: | 2014-10-18 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | GacA is essential for Group A Streptococcus and defines a new class of monomeric dTDP-4-dehydrorhamnose reductases (RmlD).
Mol.Microbiol., 98, 2015
|
|
4WGH
 
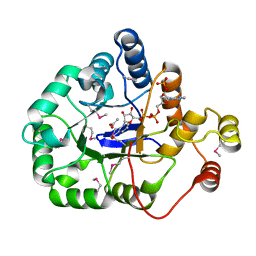 | | Crystal structure of aldo/keto reductase from Klebsiella pneumoniae in complex with NADP and acetate at 1.8 A resolution | | Descriptor: | ACETATE ION, Aldehyde reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Bacal, P, Shabalin, I.G, Cooper, D.R, Hillerich, B.S, Zimmerman, M.D, Chowdhury, S, Hammonds, J, Al Obaidi, N, Gizzi, A, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-09-18 | | Release date: | 2014-10-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of aldo/keto reductase from Klebsiella pneumoniae in complex with NADP and acetate at 1.8 A resolution
to be published
|
|
4WEV
 
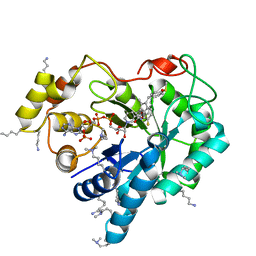 | | Crystal structure of human AKR1B10 complexed with NADP+ and sulindac | | Descriptor: | Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, [(1Z)-5-fluoro-2-methyl-1-{4-[methylsulfinyl]benzylidene}-1H-inden-3-yl]acetic acid | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Crespo, I, Porte, S, Pares, X, Farres, J, Podjarny, A. | | Deposit date: | 2014-09-11 | | Release date: | 2015-01-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.453 Å) | | Cite: | Structural analysis of sulindac as an inhibitor of aldose reductase and AKR1B10.
Chem.Biol.Interact., 234, 2015
|
|
4WF8
 
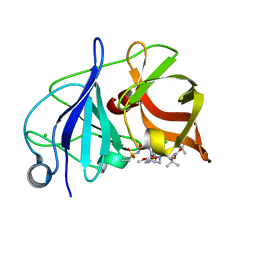 | | Crystal structure of NS3/4A protease in complex with Asunaprevir | | Descriptor: | CHLORIDE ION, N-(tert-butoxycarbonyl)-3-methyl-L-valyl-(4R)-4-[(7-chloro-4-methoxyisoquinolin-1-yl)oxy]-N-{(1R,2S)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethenylcyclopropyl}-L-prolinamide, NS3 protein, ... | | Authors: | Schiffer, C.A, Soumana, D.I, Ali, A. | | Deposit date: | 2014-09-13 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Analysis of Asunaprevir Resistance in HCV NS3/4A Protease.
Acs Chem.Biol., 9, 2014
|
|
4WEN
 
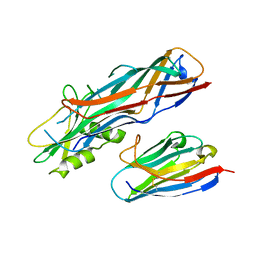 | | Co-complex structure of the F4 fimbrial adhesin FaeG variant ac with llama single domain antibody V2 | | Descriptor: | Anti-F4+ETEC bacteria VHH variable region, K88 fimbrial protein AC | | Authors: | Moonens, K, Van den Broeck, I, Pardon, E, De Kerpel, M, Remaut, H, De Greve, H. | | Deposit date: | 2014-09-10 | | Release date: | 2015-02-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural insight in the inhibition of adherence of F4 fimbriae producing enterotoxigenic Escherichia coli by llama single domain antibodies.
Vet. Res., 46, 2015
|
|
4WI3
 
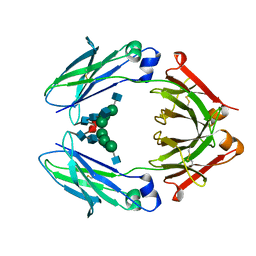 | | Structural mapping of the human IgG1 binding site for FcRn: hu3S193 Fc mutation I253A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Ig gamma-1 chain C region | | Authors: | Farrugia, W, Burvenich, I.J.G, Scott, A.M, Ramsland, P.A. | | Deposit date: | 2014-09-25 | | Release date: | 2015-09-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Structural and functional mapping of human IgG1 binding site for FcRn in vivo using human FcRn transgenic mice
To Be Published
|
|
4WI6
 
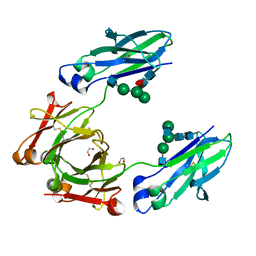 | | Structural mapping of the human IgG1 binding site for FcRn: hu3S193 Fc mutation N434A | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Ig gamma-1 chain C region | | Authors: | Farrugia, W, Burvenich, I.J.G, Scott, A.M, Ramsland, P.A. | | Deposit date: | 2014-09-25 | | Release date: | 2015-09-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structural and functional mapping of human IgG1 binding site for FcRn in vivo using human FcRn transgenic mice
To Be Published
|
|
8BQY
 
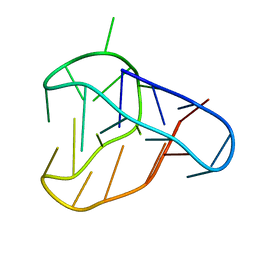 | | An i-motif domain able to undergo pH-dependent conformational transitions (acidic structure) | | Descriptor: | DNA (5'-D(*CP*(DNR)P*GP*TP*TP*(DNR)P*(DNR)P*GP*TP*TP*TP*TP*TP*CP*CP*GP*TP*TP*(DNR)P*CP*GP*T)-3') | | Authors: | Serrano-Chacon, I, Mir, B, Cupellini, L, Colizzi, F, Orozco, M, Escaja, N, Gonzalez, C. | | Deposit date: | 2022-11-22 | | Release date: | 2023-02-22 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | pH-Dependent Capping Interactions Induce Large-Scale Structural Transitions in i-Motifs.
J.Am.Chem.Soc., 145, 2023
|
|
8BV6
 
 | | An i-motif domain able to undergo pH-dependent conformational transitions (neutral structure) | | Descriptor: | DNA (5'-D(*CP*(DNR)P*GP*TP*TP*CP*(DNR)P*GP*TP*TP*TP*TP*TP*CP*CP*GP*TP*TP*CP*CP*GP*T)-3') | | Authors: | Serrano-Chacon, I, Mir, B, Cupellini, L, Colizzi, F, Orozco, M, Escaja, N, Gonzalez, C. | | Deposit date: | 2022-12-01 | | Release date: | 2023-02-22 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | pH-Dependent Capping Interactions Induce Large-Scale Structural Transitions in i-Motifs.
J.Am.Chem.Soc., 145, 2023
|
|
