1KQK
 
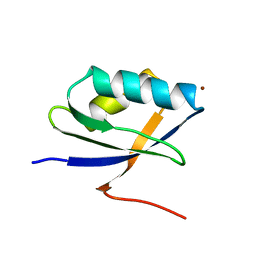 | | Solution Structure of the N-terminal Domain of a Potential Copper-translocating P-type ATPase from Bacillus subtilis in the Cu(I)loaded State | | Descriptor: | COPPER (I) ION, POTENTIAL COPPER-TRANSPORTING ATPASE | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, D'Onofrio, M, Gonnelli, L, Marhuenda-Egea, F.C, Ruiz-Duenas, F.J. | | Deposit date: | 2002-01-07 | | Release date: | 2002-04-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal domain of a potential copper-translocating P-type ATPase from Bacillus subtilis in the apo and Cu(I) loaded states.
J.Mol.Biol., 317, 2002
|
|
4QF2
 
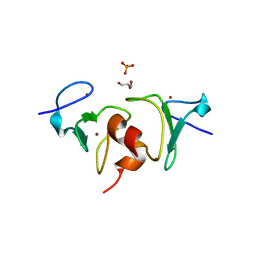 | | Crystal structure of human BAZ2A PHD zinc finger in the free form | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2A, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Tallant, C, Overvoorde, L, Van Molle, I, Chirgadze, D.Y, Ciulli, A. | | Deposit date: | 2014-05-19 | | Release date: | 2014-07-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular basis of histone tail recognition by human TIP5 PHD finger and bromodomain of the chromatin remodeling complex NoRC.
Structure, 23, 2015
|
|
6DHH
 
 | | RT XFEL structure of Photosystem II 400 microseconds after the second illumination at 2.2 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Chatterjee, R, Young, I.D, Fuller, F.D, Lassalle, L, Ibrahim, M, Gul, S, Fransson, T, Brewster, A.S, Alonso-Mori, R, Hussein, R, Zhang, M, Douthit, L, de Lichtenberg, C, Cheah, M.H, Shevela, D, Wersig, J, Seufert, I, Sokaras, D, Pastor, E, Weninger, C, Kroll, T, Sierra, R.G, Aller, P, Butryn, A, Orville, A.M, Liang, M, Batyuk, A, Koglin, J.E, Carbajo, S, Boutet, S, Moriarty, N.W, Holton, J.M, Dobbek, H, Adams, P.D, Bergmann, U, Sauter, N.K, Zouni, A, Messinger, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2018-05-20 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of the intermediates of Kok's photosynthetic water oxidation clock.
Nature, 563, 2018
|
|
6X1S
 
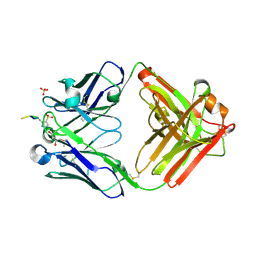 | | Structure of pHis Fab (SC1-1) in complex with pHis mimetic peptide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, NM23-1-pTza peptide, SC1-1 Heavy chain, ... | | Authors: | Kalagiri, R, Stanfield, R.L, Wilson, I.A, Hunter, T. | | Deposit date: | 2020-05-19 | | Release date: | 2021-02-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for differential recognition of phosphohistidine-containing peptides by 1-pHis and 3-pHis monoclonal antibodies.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6OIL
 
 | | Crystal structure of human VISTA extracellular domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, V-type immunoglobulin domain-containing suppressor of T-cell activation | | Authors: | Mehta, N, Cochran, J.R, Mathews, I.I. | | Deposit date: | 2019-04-09 | | Release date: | 2019-09-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and Functional Binding Epitope of V-domain Ig Suppressor of T Cell Activation.
Cell Rep, 28, 2019
|
|
6OEC
 
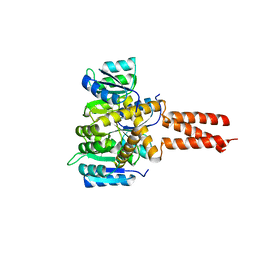 | | Yeast Spc42 Trimeric Coiled-Coil Amino Acids 181-211 fused to PDB: 3H5I | | Descriptor: | CALCIUM ION, Response regulator/sensory box protein/GGDEF domain protein,Spindle pole body component SPC42 | | Authors: | Drennan, A.C, Shivaani, K, Seeger, M.A, Andreas, M.P, Gardner, J.M, Sether, E.K.R, Jasperson, S.L, Rayment, I. | | Deposit date: | 2019-03-27 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.514 Å) | | Cite: | Structure and function of Spc42 coiled-coils in yeast centrosome assembly and duplication.
Mol.Biol.Cell, 30, 2019
|
|
7P66
 
 | | Globular glial tauopathy type 1 tau filament | | Descriptor: | Microtubule-associated protein tau | | Authors: | Shi, Y, Zhang, W, Yang, Y, Murzin, A.G, Falcon, B, Kotecha, A, van Beers, M, Tarutani, A, Kametani, F, Garringer, H.J, Vidal, R, Hallinan, G.I, Lashley, T, Saito, Y, Murayama, S, Yoshida, M, Tanaka, H, Kakita, A, Ikeuchi, T, Robinson, A.C, Mann, D.M.A, Kovacs, G.G, Revesz, T, Ghetti, B, Hasegawa, M, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2021-07-15 | | Release date: | 2021-09-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure-based classification of tauopathies.
Nature, 598, 2021
|
|
5MAU
 
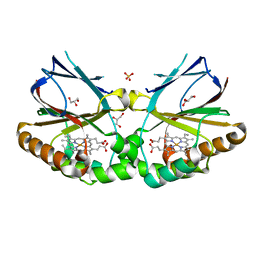 | | Crystal structure of dimeric chlorite dismutase from Cyanothece sp. PCC7425 (pH 6.5) | | Descriptor: | Chlorite dismutase, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Puehringer, D, Schaffner, I, Mlynek, G, Obinger, C, Djinovic-Carugo, K. | | Deposit date: | 2016-11-04 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Molecular Mechanism of Enzymatic Chlorite Detoxification: Insights from Structural and Kinetic Studies.
ACS Catal, 7, 2017
|
|
6DHG
 
 | | RT XFEL structure of Photosystem II 150 microseconds after the second illumination at 2.5 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Chatterjee, R, Young, I.D, Fuller, F.D, Lassalle, L, Ibrahim, M, Gul, S, Fransson, T, Brewster, A.S, Alonso-Mori, R, Hussein, R, Zhang, M, Douthit, L, de Lichtenberg, C, Cheah, M.H, Shevela, D, Wersig, J, Seufert, I, Sokaras, D, Pastor, E, Weninger, C, Kroll, T, Sierra, R.G, Aller, P, Butryn, A, Orville, A.M, Liang, M, Batyuk, A, Koglin, J.E, Carbajo, S, Boutet, S, Moriarty, N.W, Holton, J.M, Dobbek, H, Adams, P.D, Bergmann, U, Sauter, N.K, Zouni, A, Messinger, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2018-05-20 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of the intermediates of Kok's photosynthetic water oxidation clock.
Nature, 563, 2018
|
|
6OFB
 
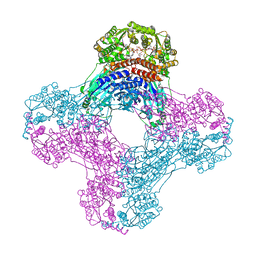 | | Crystal structure of human glutamine-dependent NAD+ synthetase complexed with NaAD+, AMP, pyrophosphate, and Mg2+ | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, Glutamine-dependent NAD(+) synthetase, ... | | Authors: | Chuenchor, W, Doukov, T.I, Gerratana, B. | | Deposit date: | 2019-03-28 | | Release date: | 2020-01-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Different ways to transport ammonia in human and Mycobacterium tuberculosis NAD+synthetases.
Nat Commun, 11, 2020
|
|
6OFM
 
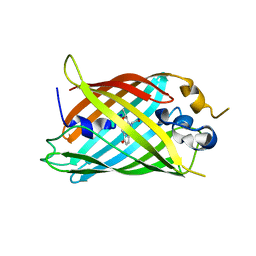 | | Crystal structure of green fluorescent protein (GFP); S65T, Y66(3-CH3Y); ih circular permutant (50-51) | | Descriptor: | Green fluorescent protein (GFP); S65T, Y66(3-CH3Y); ih circular permutant (50-51) | | Authors: | Lin, C.-Y, Romei, M.G, Mathews, I.I, Boxer, S.G. | | Deposit date: | 2019-03-31 | | Release date: | 2019-07-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Unified Model for Photophysical and Electro-Optical Properties of Green Fluorescent Proteins.
J.Am.Chem.Soc., 141, 2019
|
|
7TPG
 
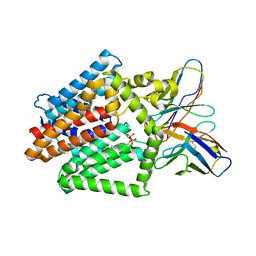 | | Single-Particle Cryo-EM Structure of the WaaL O-antigen ligase in its ligand bound state | | Descriptor: | Fab Heavy (H) Chain, Fab Light (L) Chain, GERANYL DIPHOSPHATE, ... | | Authors: | Ashraf, K.U, Nygaard, R, Vickery, O.N, Erramilli, S.K, Herrera, C.M, McConville, T.H, Petrou, V.I, Giacometti, S.I, Dufrisne, M.B, Nosol, K, Zinkle, A.P, Graham, C.L.B, Loukeris, M, Kloss, B, Skorupinska-Tudek, K, Swiezewska, E, Roper, D, Clarke, O.B, Uhlemann, A.C, Kossiakoff, A.A, Trent, M.S, Stansfeld, P.J, Mancia, F. | | Deposit date: | 2022-01-25 | | Release date: | 2022-04-06 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural basis of lipopolysaccharide maturation by the O-antigen ligase.
Nature, 604, 2022
|
|
7TPJ
 
 | | Single-Particle Cryo-EM Structure of the WaaL O-antigen ligase in its apo state | | Descriptor: | Fab Heavy (H) Chain, Fab Light (L) Chain, Putative cell surface polysaccharide polymerase/ligase | | Authors: | Ashraf, K.U, Nygaard, R, Vickery, O.N, Erramilli, S.K, Herrera, C.M, McConville, T.H, Petrou, V.I, Giacometti, S.I, Dufrisne, M.B, Nosol, K, Zinkle, A.P, Graham, C.L.B, Loukeris, M, Kloss, B, Skorupinska-Tudek, K, Swiezewska, E, Roper, D, Clarke, O.B, Uhlemann, A.C, Kossiakoff, A.A, Trent, M.S, Stansfeld, P.J, Mancia, F. | | Deposit date: | 2022-01-25 | | Release date: | 2022-04-06 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structural basis of lipopolysaccharide maturation by the O-antigen ligase.
Nature, 604, 2022
|
|
5M7O
 
 | | Crystal structure of NtrX from Brucella abortus processed with the CrystalDirect automated mounting and cryo-cooling technology | | Descriptor: | MAGNESIUM ION, Nitrogen assimilation regulatory protein | | Authors: | Cornaciu, I, Fernandez, I, Hoffmann, G, Carrica, M.C, Goldbaum, F.A, Marquez, J.A. | | Deposit date: | 2016-10-28 | | Release date: | 2017-01-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-Dimensional Structure of Full-Length NtrX, an Unusual Member of the NtrC Family of Response Regulators.
J. Mol. Biol., 429, 2017
|
|
6O9M
 
 | | Structure of the human apo TFIIH | | Descriptor: | CDK-activating kinase assembly factor MAT1, General transcription factor IIH subunit 1, General transcription factor IIH subunit 2, ... | | Authors: | Yan, C.L, Dodd, T, He, Y, Tainer, J.A, Tsutakawa, S.E, Ivanov, I. | | Deposit date: | 2019-03-14 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Transcription preinitiation complex structure and dynamics provide insight into genetic diseases.
Nat.Struct.Mol.Biol., 26, 2019
|
|
5C44
 
 | | Crystal structure of a transcribing RNA Polymerase II complex reveals a complete transcription bubble | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Barnes, C.O, Calero, M, Malik, I, Spahr, H, Zhang, Q, Pullara, F, Kaplan, C.D, Calero, G. | | Deposit date: | 2015-06-17 | | Release date: | 2015-08-26 | | Method: | X-RAY DIFFRACTION (3.95 Å) | | Cite: | Crystal Structure of a Transcribing RNA Polymerase II Complex Reveals a Complete Transcription Bubble.
Mol.Cell, 59, 2015
|
|
5ME0
 
 | | Structure of the 30S Pre-Initiation Complex 1 (30S IC-1) Stalled by GE81112 | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Lopez-Alonso, J.P, Fabbretti, A, Kaminishi, T, Iturrioz, I, Brandi, L, Gil Carton, D, Gualerzi, C, Fucini, P, Connell, S. | | Deposit date: | 2016-11-14 | | Release date: | 2017-01-11 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (13.5 Å) | | Cite: | Structure of a 30S pre-initiation complex stalled by GE81112 reveals structural parallels in bacterial and eukaryotic protein synthesis initiation pathways.
Nucleic Acids Res., 45, 2017
|
|
7U2E
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody ADI-55688 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADI-55688 heavy chain, ADI-55688 light chain, ... | | Authors: | Yuan, M, Zhu, X, Wilson, I.A. | | Deposit date: | 2022-02-23 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A broad and potent neutralization epitope in SARS-related coronaviruses.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7U2D
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody ADG20 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADG20 heavy chain, ADG20 light chain, ... | | Authors: | Zhu, X, Yuan, M, Wilson, I.A. | | Deposit date: | 2022-02-23 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | A broad and potent neutralization epitope in SARS-related coronaviruses.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5C4J
 
 | | Crystal structure of a transcribing RNA Polymerase II complex reveals a complete transcription bubble | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Barnes, C.O, Calero, M, Malik, I, Spahr, H, Zhang, Q, Pullara, F, Kaplan, C.D, Calero, G. | | Deposit date: | 2015-06-18 | | Release date: | 2015-08-26 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Crystal Structure of a Transcribing RNA Polymerase II Complex Reveals a Complete Transcription Bubble.
Mol.Cell, 59, 2015
|
|
6OFK
 
 | | Crystal structure of green fluorescent protein (GFP); S65T; ih circular permutant (50-51) | | Descriptor: | ACETATE ION, Green Fluorescent Protein (GFP); S65T; ih circular permutant (50-51) | | Authors: | Lin, C.-Y, Romei, M.G, Mathews, I.I, Boxer, S.G. | | Deposit date: | 2019-03-30 | | Release date: | 2019-07-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Unified Model for Photophysical and Electro-Optical Properties of Green Fluorescent Proteins.
J.Am.Chem.Soc., 141, 2019
|
|
5ME1
 
 | | Structure of the 30S Pre-Initiation Complex 2 (30S IC-2) Stalled by GE81112 | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Lopez-Alonso, J.P, Fabbretti, A, Kaminishi, T, Iturrioz, I, Brandi, L, Gil Carton, D, Gualerzi, C, Fucini, P, Connell, S. | | Deposit date: | 2016-11-14 | | Release date: | 2017-01-11 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (13.5 Å) | | Cite: | Structure of a 30S pre-initiation complex stalled by GE81112 reveals structural parallels in bacterial and eukaryotic protein synthesis initiation pathways.
Nucleic Acids Res., 45, 2017
|
|
7OMT
 
 | | Crystal structure of ProMacrobody 21 with bound maltose | | Descriptor: | HEXAETHYLENE GLYCOL, MAGNESIUM ION, ProMacrobody 21, ... | | Authors: | Botte, M, Ni, D, Schenck, S, Zimmermann, I, Chami, M, Bocquet, N, Egloff, P, Bucher, D, Trabuco, M, Cheng, R.K.Y, Brunner, J.D, Seeger, M.A, Stahlberg, H, Hennig, M. | | Deposit date: | 2021-05-24 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cryo-EM structures of a LptDE transporter in complex with Pro-macrobodies offer insight into lipopolysaccharide translocation.
Nat Commun, 13, 2022
|
|
5WOW
 
 | |
5M7N
 
 | | Crystal structure of NtrX from Brucella abortus in complex with ATP processed with the CrystalDirect automated mounting and cryo-cooling technology | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Nitrogen assimilation regulatory protein | | Authors: | Cornaciu, I, Fernandez, I, Hoffmann, G, Carrica, M.C, Goldbaum, F.A, Marquez, J.A. | | Deposit date: | 2016-10-28 | | Release date: | 2017-01-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Three-Dimensional Structure of Full-Length NtrX, an Unusual Member of the NtrC Family of Response Regulators.
J. Mol. Biol., 429, 2017
|
|
